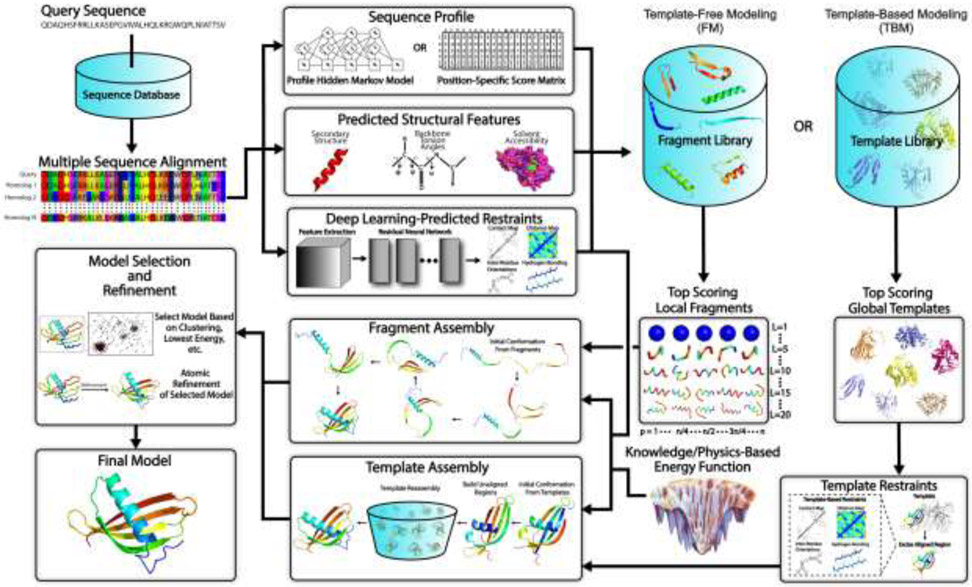
Fig 1.
Typical steps involved in template-free and template-based protein structure prediction approaches. Starting from a query sequence, an MSA is generated by identifying homologous sequences from a sequence database. The MSA is then converted into a sequence profile and used to predict structural features such as the secondary structure, backbone torsion angles and solvent accessibility. For fragment assembly-based FM methods, these structural features together with the sequence profile are used to search a fragment library to identify high scoring local fragments. For TBM methods, they are used by threading protocols to identify global template structures. Meanwhile, co-evolutionary information is extracted from the MSA and fed into a deep residual neural network to predict spatial restraints such as inter-residue long-range contacts, distances, hydrogen bonds and torsion angles. For full-length model construction, structure assembly simulations are performed under the guidance of a composite force field which usually combines the generic knowledge- and/or physics-based energy function with deep neural network feature prediction (plus template-based restraints in the case of TBM). Finally, representative models are typically selected from the lowest energy conformations or based on structural clustering, followed by atomic-level refinement to generate the final model.