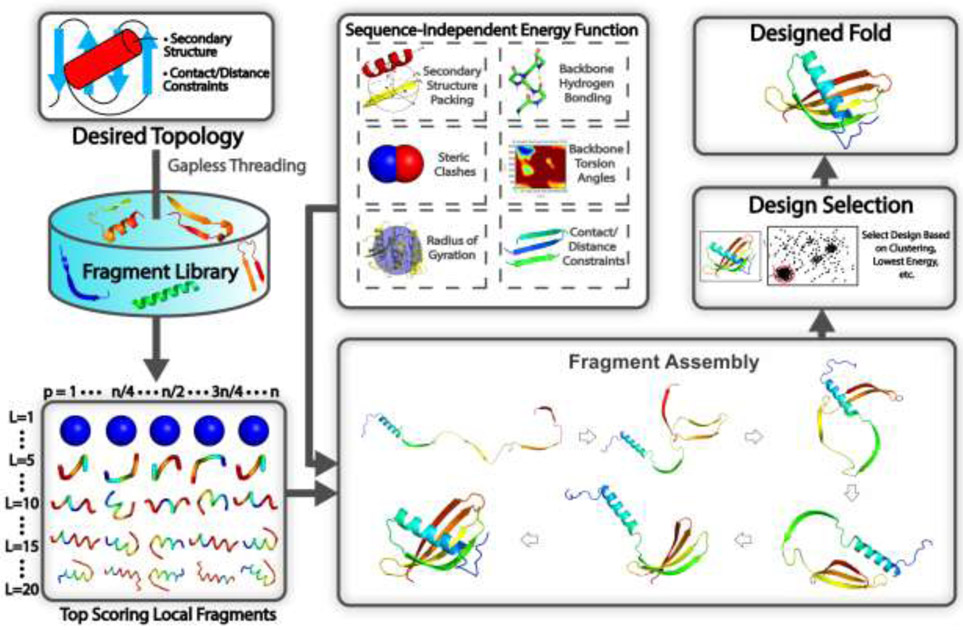
Fig 3.
Typical steps involved in a fragment assembly-based approach to design new protein structures. Starting from the desired secondary structure together with user-defined packing restraints, such as residue-residue contact/distance restraints, the query is searched through a non-redundant PDB structure library using gapless threading to generate position-specific fragment structures. High scoring fragments, which may range from 1-20 residues long, are identified based on the complementarity between the desired secondary structure and a fragment’s secondary structure and backbone torsion angles. Then during the folding simulations, the top scoring local fragments are assembled under the guidance of a sequence-independent energy function, which accounts for fundamental rules that govern protein folding such as secondary structure packing, backbone hydrogen bonding, favorable backbone torsion angles, steric clashes, radius of gyration, as well as the artificial contact/distance restraints supplied by the user. As the method is sequence independent, generic side-chain centers of mass, typically those for valine, are used to evaluate energy terms such as steric clashes. Following the folding simulations, the final design may be selected based on clustering of the simulation decoys, by selecting the lowest energy structure, or through whatever filter the user deems appropriate.