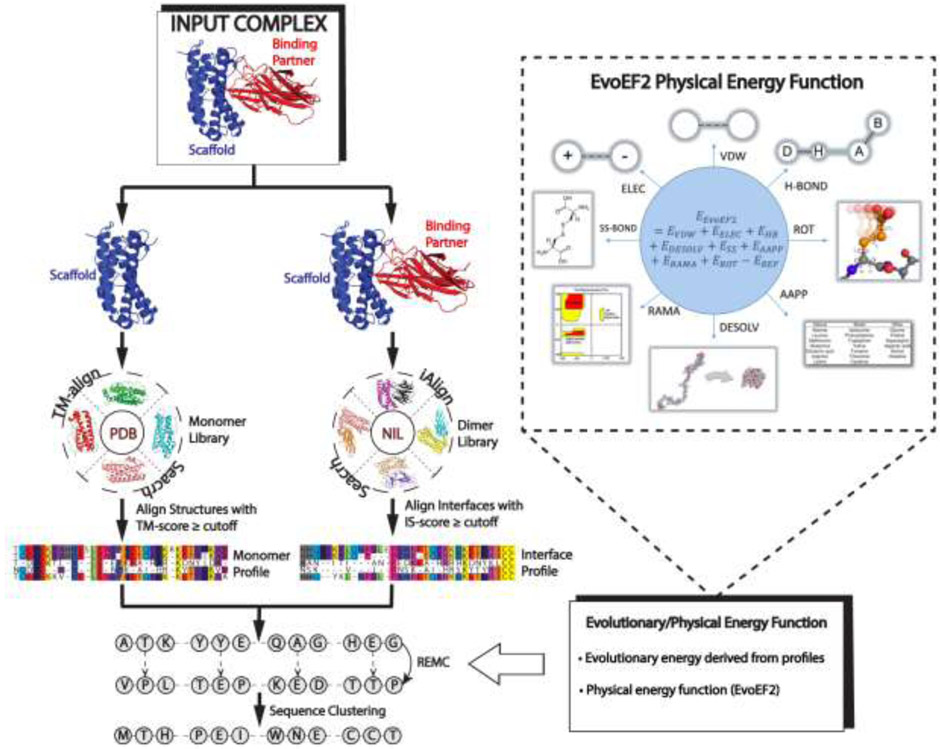
Fig 4.
A protocol for evolution-based protein-protein interaction design used by EvoDesign. The procedure starts from an input complex, for which monomer/interface structural homologs are identified from the PDB library through TM-align and iAlign searches, respectively. Structural profiles are then constructed from the alignments of the monomer/interface analogs and used in conjunction with a physics-based potential, EvoEF2, to guide the REMC simulations to design novel protein sequences. The final designs are selected from the center of the largest cluster of designed sequence decoys.