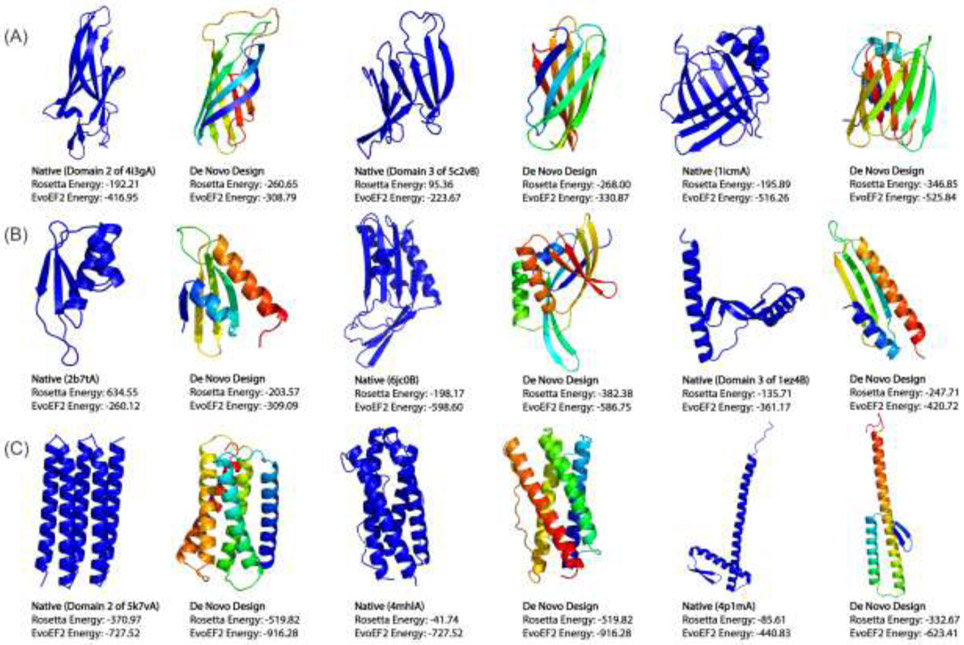
Fig 5.
Protein folds designed de novo starting from 9 unique secondary structures. The designed folds and corresponding wildtype native proteins (with denoted PDB IDs) whose secondary structures were used as input are shown side-by-side for (A) 3 β proteins, (B) 3 α/β and α+β proteins, and (C) 3 a proteins. Even in the absence of pre-defined packing rules, such as inter-residue distance restraints, the designed new folds have well-packed topologies with lower or comparable Rosetta and EvoEF2 energies.