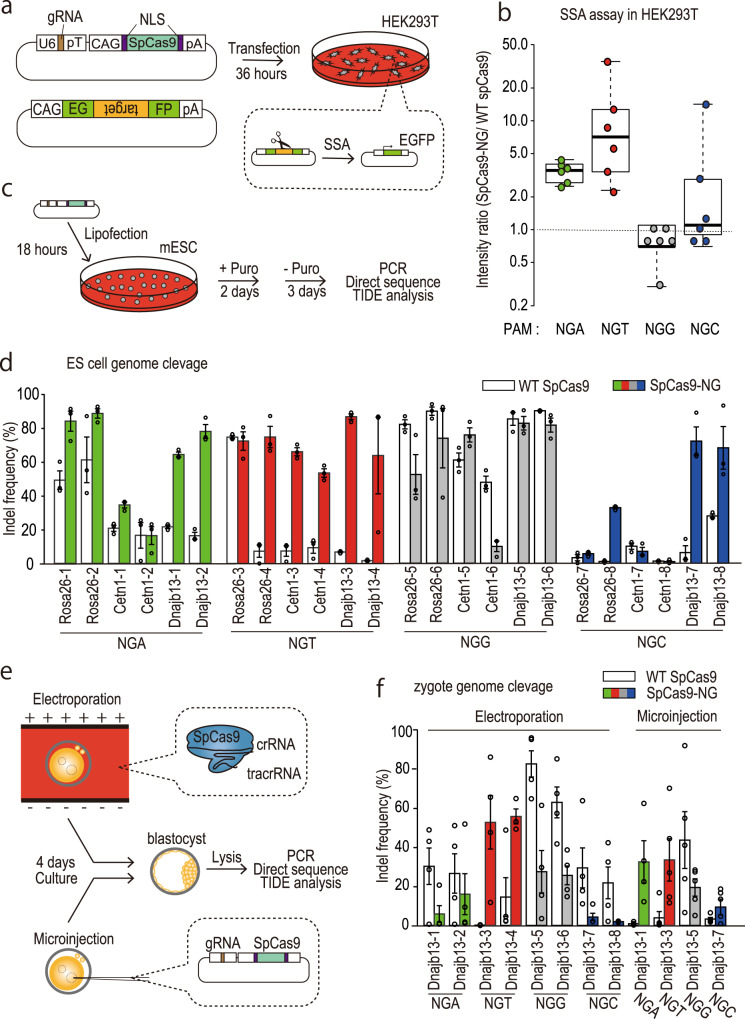
Fig. 1. Genome editing efficiency using SpCas9-NG.
a Strategy of the single-strand annealing (SSA) assay used to evaluate DNA cleavage activity. After transfection of 293 T cells with gRNA/Cas9 and reporter plasmids, the EGFP fluorescence intensity was subsequently measured. b Intensity ratio (SpCas9-NG /WT-SpCas9) in the SSA assay. c Strategy of ESC genome editing for examining indel efficiency. After the transfection of ES cells with gRNA/Cas9 plasmids targeting Rosa26, Cetn1, and Dnajb13 loci, we examined indel efficiency by directly sequencing the PCR amplicons and TIDE software. d Indel frequency in pooled ES cells determined by TIDE analysis (n = 3, taken from distinct samples). Error bars indicate standard error. e Strategy of zygote genome editing for examining indel efficiency. After electroporating gRNA/SpCas9 ribonucleoprotein complexes or microinjecting gRNA/SpCas9 plasmids targeting the Dnajb13 locus into mouse zygotes, we examined indel efficiency in blastocysts by direct sequencing of the PCR amplicons and TIDE software. f Indel frequency in pooled blastocysts was determined by TIDE analysis (n = 3; taken from distinct samples). Error bars indicate standard error.