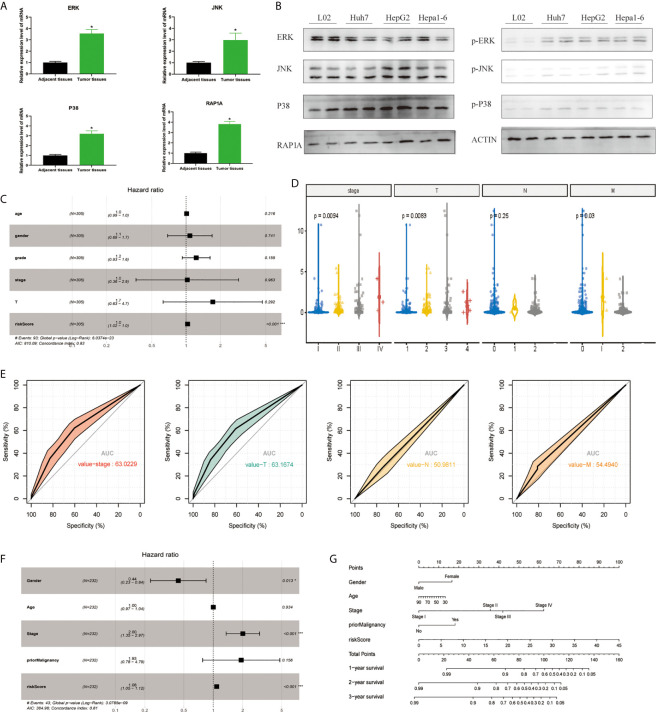
Figure 1.
Establishment of the risk signature with MAPK-RAP1A related genes in the training (TCGA) and validation database (ICGC). (A) RT-qPCR was used to detect the expression of ERK, JNK, p38, RAP1A in 20 cases HCC tumor tissues and adjacent tissues. (B) Western blot was used to detect the expression of ERK, JNK, p38, RAP1A in Huh7, HepG2, Hepa1-6, and L02 cell lines. (C) Multivariate Cox regression analysis to validate prognosis related clinicopathological characters. (D) The correlation of our MAPK-RAP1A risk signature with the clinicopathological characters (TNM stage) of HCC from the training set. (E) ROC curves of AUC evaluated the efficiency of the risk signature for predicting TNM stage in training set. (F) Multivariate Cox regression analysis to validate prognosis related clinicopathological characters. (G) Nomogram for predicting 1-, 3-, and 5-year OS of LIHC patients in the ICGC cohort. *p < 0.05; ***p < 0.001.