FIGURE 3.
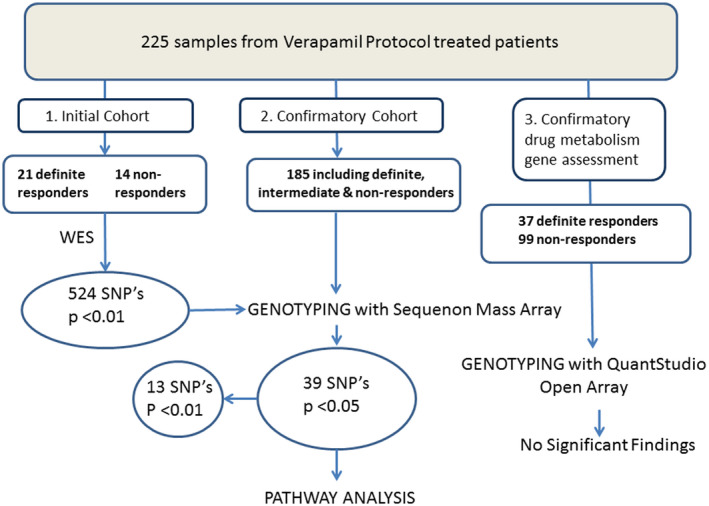
Schematic summary of the study procedures: Whole‐exome sequencing was carried out in 21 definite responders and 14 definite non‐responders to verapamil and 524 SNPs were found to have a significant association with treatment response with p‐value < 0.01. The 524 identified SNP’s were then genotyped using Sequenom custom panel approach in much larger, separate cohort of 185 patients. After genotyping 39 SNP’s attained significance of p < 0.05 and of those 13 reached of p‐value <0.01. Ingenuity pathway analysis was then performed using SNP’s with p < 0.05 and Ingenuity Protein network analysis was carried out using the SNP’s with p < 0.01. To assess whether the observed treatment response to verapamil might be related to variation in the genes involved in verapamil metabolism or cellular transport, all the definite responders and non‐responders from the combined WES and larger Sequenom MassARRAY genotyped cohorts (37 definite responders and 99 definite non‐responders) were genotyped for variants in CYP2C8, CYP3A4 and CYP3A5 as well as in the transporter gene ABCB1 (c.3435T>C, count C; rs2032582 c.2677G>T/A, count A; c.2677G>T/A, count T) with QuantStudio OpenArray
