Abstract
Increasing evidence has indicated that long non-coding RNAs (lncRNAs) serve an essential role in carcinogenesis and cancer development. It has been reported that lncRNA nicotinamide nucleotide transhydrogenase antisense RNA 1 (NNT-AS1) serves a crucial role in several types of cancer. However, the clinical significance of circulating NNT-AS1 expression in colorectal cancer (CRC) remains to be elucidated. The current study aimed to investigate the potential role of NNT-AS1 and the clinical significance of its serum expression levels in patients with CRC. The expression of NNT-AS1 was measured in 40 pairs of tumor and adjacent normal tissues from patients with CRC via reverse transcription-quantitative PCR. The serum expression levels of NNT-AS1 were assayed in an independent cohort of healthy controls and patients with CRC. The levels of NNT-AS1 were also compared between paired preoperative and postoperative serum samples. In addition, the presence of exosomal NNT-AS1 in serum was explored. Furthermore, the biological roles of NNT-AS1 were investigated in CRC cells in vitro. The expression of NNT-AS1 was significantly upregulated in tumor tissues compared with adjacent normal tissues (P<0.05). A higher level of NNT-AS1 was associated with an advanced CRC stage. The serum levels of NNT-AS1 were significantly upregulated in patients with CRC compared with healthy subjects (P<0.05). Furthermore, the NNT-AS1 levels were significantly decreased in postoperative samples compared with preoperative samples (P<0.01). In addition, it was also identified that NNT-AS1 was upregulated in CRC exosomes (P<0.01), whereas no significant difference was observed in NNT-AS1 levels between serum and exosomes. Silencing of NNT-AS1 inhibited the proliferation, migration and invasion of CRC cells. It was also identified that NNT-AS1 exerted its effects via regulation of the microRNA-496/Ras-related protein Rap-2c axis. The present study demonstrated that circulating NNT-AS1, which may be protected by exosomes, could be a novel potential biomarker and therapeutic target in CRC.
Keywords: nicotinamide nucleotide transhydrogenase antisense RNA 1, exosome, colorectal cancer, microRNA-496, Ras-related protein Rap-2c, biomarker
Introduction
Colorectal cancer (CRC) is a primary cause of cancer-related deaths, with >1.2 million newly diagnosed cases every year worldwide (1). Although great achievements have been made in diagnostic approaches and therapeutic strategies, the clinical outcome of advanced CRC remains poor (2). It has been identified that the survival rate of patients with CRC is highly associated with the stage, with ~90% for stage I and only 25% for stage II (3). Therefore, there is an urgent requirement to identify reliable biomarkers of CRC progression and provide potential therapeutic targets.
Long non-coding (lnc)RNAs are a group of non-coding (nc)RNAs with a length of >200 nucleotides (4). For a long time, lncRNAs were considered the ‘junk’ of the genome due to their inability to encode proteins (5). In previous years, accumulating evidence has demonstrated that lncRNAs serve essential roles in the initiation and progression of cancer, and can therefore be used as reliable diagnostic and prognostic biomarkers (6). Notably, lncRNAs are relatively stable and it is feasible to detect cancer-related lncRNAs in blood and/or other body fluids (7). lncRNA nicotinamide nucleotide transhydrogenase antisense RNA 1 (NNT-AS1) is a newly-identified lncRNA that is located in the chromosome 5p12 region, with a full length of 3,304 bp (8). NNT-AS1 is abnormally expressed in various types of cancer, including cervical cancer, osteosarcoma, gastric cancer and lung cancer (9–11). NNT-AS1 is also identified to be highly upregulated in CRC tissues compared with adjacent normal tissues and can contribute to the migration and invasion of CRC cells (12). However, the clinical significance of circulating NNT-AS1 expression in CRC has not yet been investigated.
Previous studies have identified that nanosized vesicles, also known as exosomes, may function as natural carriers to transport proteins, mRNAs and various ncRNAs (13,14). Exosomes can be identified in various human body fluids, such as plasma, saliva, serum and urine (14). Their double layer construction makes exosomes ideal vehicles that protect biomolecules from lysosomal and/or other enzymes. Exosomes are involved not only in cell-to-cell communication, but also in the progression of tumorigenesis (15). Certain lncRNAs can be selectively packaged into exosomes, which are correlated with the clinicopathological features of cancer, and therefore may have useful applications as cancer-related biomarkers (16).
MicroRNAs (miRNAs/miRs) are another type of ncRNA with a length of ~20 nucleotides. A number of studies have identified that miRNAs are involved in the pathogenesis of various human diseases, including types of human cancers (17). Among such miRNAs, miR-496 has been identified as a regulator in the progression of different cancers, including lung cancer, breast cancer and osteosarcoma (18,19). However, there are few studies on the role of miR-496 in CRC. Ras-related protein Rap-2c (RAP2C) belongs to the GTP-binding protein family and has been identified to participate in the development of different types of cancer. For instance, RAP2C promotes the migration and invasion of osteosarcoma cells (20). In addition, RAP2C has also been identified to promote the tumorigenesis of breast cancer and laryngeal squamous cell carcinoma (21,22). However, there remains a lack of studies regarding the role of RAP2C in CRC.
The current study evaluated the expression levels of NNT-AS1 in CRC tissues and adjacent normal tissues, followed by analysis of serum NNT-AS expression in patients with CRC and healthy controls. The presence of serum NNT-AS1 in exosomes was also determined and it indicated that exosomal NNT-AS1 may be a novel biomarker for CRC. In addition, the role of NNT-AS1 in CRC was also investigated and it was demonstrated that NNT-AS1 exerted its effects on CRC cells via regulation of the miR-496/RAP2C axis.
Materials and methods
Patient and healthy control specimens
The present study was approved by the Ethical Committee of Hwamei Hospital, University of Chinese Academy of Sciences (Ningbo, China; approval no. 43321764) and all procedures were conducted according to the Declaration of Helsinki. Written informed consent was obtained from each patient. All patients and healthy controls involved in this study were recruited from Hwamei Hospital between January 2017 and August 2019 according to the following criteria: i) Patients who underwent surgery with curative intent; ii) relevant clinical features (Table I) are available; iii) patients who did not receive radiotherapy or chemotherapy; iv) patients without other tumors. A total of 40 resected tumor tissues and adjacent normal tissues (≥1 cm from tumor tissue) were obtained at the Department of General Surgery. The diagnosis of CRC was histopathologically confirmed and other clinical data, including age, sex, tumor location, tumor size, differentiation, lymph node metastasis and distant metastasis, were collected. The postoperative pathological staging was evaluated based on the 7th edition of the Union for International Cancer Control tumor-node-metastasis (TNM) staging system (23). Serum samples were obtained from 40 age- and sex-matched healthy volunteers. All patients underwent physical examinations at Hwamei Hospital before recruitment. All resected tissue samples were immediately snap-frozen in liquid nitrogen. Venous blood was collected and prepared as previously described and stored at −80°C until analysis (14).
Table I.
Relationship between NNT-AS1 expression and clinical characteristics of patients with CRC.
| Tissue expression of NNT-AS1 | Serum expression of NNT-AS1 | ||||||
|---|---|---|---|---|---|---|---|
| Characteristics | n | High | Low | P-value | High | Low | P-value |
| Sex | 0.371 | 0.446 | |||||
| Female | 14 | 6 | 8 | 7 | 7 | ||
| Male | 26 | 14 | 12 | 11 | 15 | ||
| Age, years | 0.292 | 0.503 | |||||
| ≤60 | 10 | 4 | 6 | 4 | 6 | ||
| >60 | 30 | 17 | 13 | 14 | 16 | ||
| Tumor location | 0.578 | 0.252 | |||||
| Colon | 18 | 7 | 11 | 11 | 7 | ||
| Rectum | 22 | 9 | 13 | 10 | 12 | ||
| Tumor size, cm | 0.184 | 0.564 | |||||
| ≤4 | 25 | 10 | 15 | 11 | 14 | ||
| >4 | 15 | 9 | 6 | 7 | 8 | ||
| TNM stage | 0.033 | 0.576 | |||||
| I–II | 23 | 7 | 16 | 9 | 14 | ||
| III–IV | 17 | 11 | 6 | 7 | 10 | ||
| Local invasion | 0.025 | 0.252 | |||||
| T1-T2 | 18 | 5 | 13 | 11 | 7 | ||
| T3-T4 | 22 | 14 | 8 | 10 | 12 | ||
| Lymph node | 0.004 | 0.501 | |||||
| No | 21 | 7 | 14 | 11 | 10 | ||
| Yes | 19 | 15 | 4 | 9 | 10 | ||
NNT-AS1, lncRNA nicotinamide nucleotide transhydrogenase antisense RNA 1; CRC, colorectal cancer; TNM, tumor-node-metastasis.
Serum exosome isolation
Exosomes were isolated and purified from 0.5 ml serum using a Total Exosome Isolation Kit (cat. no. 4484450, Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The exosomes were then suspended in 1X phosphate-buffered saline (PBS) and stored at −80°C before use.
Cell culture and transfection
Human normal colorectal epithelial cells (HCnEpC) and human CRC cells (LoVo, RKO, SW48 and HCT116) were purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. All cells were cultured at a density of 1×106 cells/ml in RPMI-1640 medium (HyClone; Cytiva) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin (Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2. For transfection, small interfering (si)RNA against NNT-AS1 (si-NNT-AS1, 5′-GAAAAGAAAAAGAAGCUUATT-3′), scramble siRNA negative control (si-NC, 5′-UUCUCCGAACGUGUCACG-3′), pcDNA3.1 vector (empty control), NNT-AS1 overexpression vector (pcDNA. NNT-AS1), miR-496 mimic (5′-UGAGUAUUACAUGGCCAAUCUC-3′), scramble negative control miRNA mimic (miR-NC) (5′-ACCGUACACGUUACCGUCACU-3′), miR-496 inhibitor (5′-GAGAUUGGCCAUGUGUAAUACUCA-3′), miR-NC inhibitor (5′-CAUAACGACUACGGUACCAUCA-3′) and RAP2C overexpression vector (pcDNA. RAP2C) were all synthesized by Shanghai GenePharma Co., Ltd. All transfections were conducted using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The working concentration of siRNA was 20 nM and that for miRNA mimics and inhibitors was 50 nM. The concentration used for plasmids was 100 nM. At 24 h after the transfection, following assays were conducted.
Total RNA purification and reverse transcription-quantitative (RT-q)PCR
Total RNA was purified from tissues (5 g)/serum (5 ml) using TRIzol® reagent (Thermo Fisher Scientific, Inc.) and exosomal RNA of serum was extracted using the mirVana™ PARIS™ RNA and Native Protein Purification Kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The quantity and quality of RNA were measured using a NanoDrop™ 2000 Spectrophotometer (Thermo Fisher Scientific, Inc.). cDNA was generated by reverse transcription using a PrimeScript RT Reagent Kit (Takara Biotechnology Co., Ltd.) according to the manufacturer's instructions. SYBR® Premix Ex Taq™ II (Takara Biotechnology Co., Ltd.) was used to perform RT-qPCR and reactions were run in duplicate at 95°C for 30 sec for 1 cycle, followed by 40 cycles of 95°C for 3 sec and 60°C for 30 sec. The total reaction volume was 50 µl. Primers were designed as follows: NNT-AS1, forward 5′-ACGTGCAGACAACATCTACCT-3′ and reverse 5′-TACAACACCTTCCCGCAT-3′; miR-496, forward 5′-GCAATCCTGAGGAAGAGGATGTGGA-3′ and reverse 5′-TCTGAGCTGCGAGTCATT-3′; GAPDH, forward 5′-CCCACTTGAAGGGTGGAGCCAA-3′ and reverse 5′-TGGCATGGACTGTGGTCATGA-3′; Tubulin, forward 5′-ATGCGTGAAATAGTTCATATCC-3′ and reverse 5′-TTAGATTTCTTCTTCCTCGTC-3′; and U6, forward 5′-GCTTCACGAATTTGCGTGTCAT-3′ and reverse 5′-GCTTCGGCAGCACATATACTAAAAT-3′. The expression of lncRNA NNT-AS1 was expressed as the 2−ΔΔCq value, with GAPDH and Tubulin as internal controls for NNT-AS1 and U6 as the internal control for miR-496 (24).
NanoSight assay flow cytometry analysis
Exosome pellets were resuspended in 1 ml PBS and the particle size distribution was analyzed under a NanoSight NS300 (Malvern Instruments, Ltd).
Flow cytometry analysis of exosomes
For the analysis of exosomes by fluorescence-activated cell sorting, the exosomes were mixed with 100 µl PBS at room temperature for 5 min and then analyzed with monoclonal antibodies against CD9 (1:200; cat. no. ab2215; Abcam), CD63 (1:200; cat. no. ab193349; Abcam) and CD81 (1:200; cat. no. ab219209; Abcam) by flow cytometry (BD FACSCalibur; BD Biosciences). The results were analyzed with FlowJo 11.0 (TreeStar Ltd.).
Cell viability assay
Cell viability was measured using an MTT assay as described previously (24). Briefly, cells were seeded in 6-well plates at a density of 1×105 cells/well. Following transfection, 20 µl of 0.5 mg/ml MTT (Sigma-Aldrich; Merck KGaA) was added to the medium and incubated for another 4 h at 37°C. Before measurement, 150 µl DMSO was added and a microplate reader (BioTek Instruments, Inc.) was used to measure the optical density value at 450 nm.
Dual-luciferase assays
The wild-type (wt) and mutant (mut) fragments of NNT-AS1 and the 3′ untranslated region of RAP2C containing the miR-496 targeted sequences were synthesized by Shanghai GenePharma Co., Ltd. and inserted into the pGL3 promoter vector (Promega Corporation). The transfection was conducted using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) according to the manufacturer's guide. The concentration of plasmid was 100 ng. Then, 48 h after transfection, cells were collected and the Dual-Luciferase Reporter Assay System (Promega Corporation) was used to measure luminescence. Renilla luciferase activity was used to normalize the firefly luciferase activity.
Cell migration and invasion assay
The wound healing assay was performed using 6-well plates (Corning Life Sciences). A total of 1×106 cells were seeded into each well and cultured with 5% serum (Gibco; Thermo Fisher Scientific, Inc.) medium overnight until 90% confluent. A 200-µl pipette tip was used to make a straight scratch. The floating cells were gently washed off using PBS and fresh full medium was added. The initial gap length (0 h) and the residual gap length at 24 h after wounding were calculated from photomicrographs as previously described (25). The results were observed using the IX70 inverted light microscope (magnification, ×100; Olympus Corporation). For the cell invasion assay, a Transwell chamber (Corning Life Sciences) was used. The upper chamber was coated with Matrigel at room temperature for 4 h (BD Biosciences) for filtering, and culture medium containing 20% FBS (Gibco; Thermo Fisher Scientific, Inc.) was added to the lower chamber. Then, 1×105 glioma cells were cultured in 2 ml serum-free medium and placed in the upper chamber. After 48 h at 37°C, non-invasive cells were removed and invasive cells were fixed with 70% ethanol at room temperature for 0.5 h and stained with 5% crystal violet (Beyotime Institute of Biotechnology) at room temperature for 0.5 h. The number of invading cells to the lower chamber was counted under the IX70 inverted optical microscope (magnification, ×100; Olympus Corporation); five fields were randomly selected from each sample. This experiment was repeated three times.
Western blot analysis
Total protein was collected with RIPA buffer (Beyotime Institute of Biotechnology) and the concentration of protein was measured using a BCA protein assay kit (Beyotime Institute of Biotechnology). Protein (20 µg) was separated via SDS-PAGE on a 10% gel, and then transferred to a PVDF membrane (EMD Millipore). After blocking with 5% skimmed milk for 1 h at room temperature, the membranes were incubated with primary antibodies overnight at 4°C. The following primary antibodies were used: Anti-CD63 (1:1,000; cat. no. ab193349; Abcam), anti-tumor susceptibility gene 101 protein (1:1,000; cat. no. ab125011; Abcam), anti-CD9 (1:1,000; cat. no. ab92726; Abcam); and anti-Flotillin-1 (1:1,000; cat. no. ab41927; Abcam). Anti-RAP2C (1:1,000; cat. no. ab97805; Abcam), GAPDH (1:5,000; cat. no. ab9485; Abcam). Subsequently, the membrane was washed three times with TBS with 1% Tween-20 for 10 min, and then incubated with goat anti-rabbit HRP-conjugated secondary antibody (1:5,000; cat. no. ab6721; Abcam) for 1 h at room temperature. Then, the membrane was visualized with enhanced chemiluminescent solution (Beijing Solarbio Science & Technology Co., Ltd.). The results were visualized using the ChemiDoc XRS+ GelDoc™ 5.0 (BioRad Laboratories, Inc.).
Bioinformatical analysis
Prediction of the NNT-AS1-miRNA-target gene was performed using the StarBase 3.0 (starbase.sysu.edu.cn) and TargetScan 2.0 (https://targetscan.org/).
Statistical analysis
Statistical analyses were performed using SPSS 22.0 (IBM Corp.) and GraphPad Prism 6.0 (GraphPad Software, Inc.). Comparisons of continuous outcomes between groups were performed using a two-tailed, paired Student's t-test and multiple groups were analyzed with one-way analysis of variance (ANOVA), followed by Tukey's post hoc test. The receiver operating characteristic (ROC) curves and area under the curve (AUC) values were calculated using algorithms described previously (26). P<0.05 was considered to indicate a statistically significant difference.
Results
NNT-AS1 is upregulated in CRC tissues
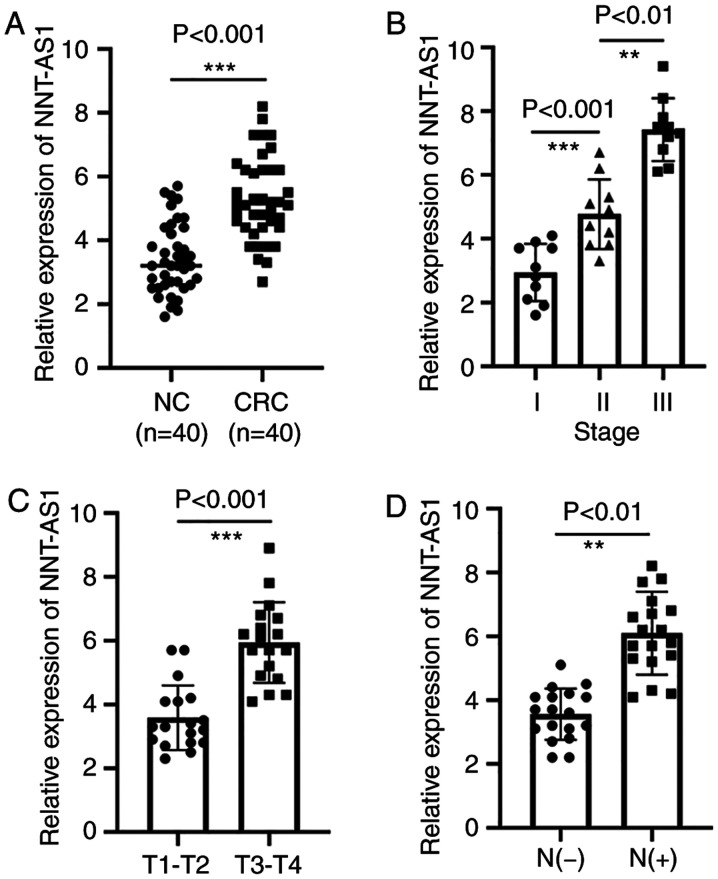
The expression levels of NNT-AS1 were measured in a set of 40 paired tissues by RT-qPCR. The expression of NNT-AS1 in tumor tissues was significantly higher than that in adjacent normal tissues (P<0.001, Fig. 1A). The association of NNT-AS1 levels with several clinicopathological features was then examined (Table I). It was identified that patients with CRC at advanced stages had higher NNT-AS1 levels (P<0.01, Fig. 1B). It was also identified that the level of NNT-AS1 was significantly associated with local invasion (P<0.001, Fig. 1C). In addition, high expression of NNT-AS1 was associated with lymph node metastasis (P<0.001, Fig. 1D). The expression of NNT-AS1 was not related to gender, age, tumor size or tumor location (P>0.05, Table I).
Figure 1.
Expression of NNT-AS1 is upregulated in CRC tissues. (A) The expression levels of NNT-AS1 were measured by reverse transcription-quantitative PCR in 40 paired CRC tissues and adjacent normal tissues. (B) The expression levels of NNT-AS1 at different tumor-node-metastasis stages of CRC. (C) Higher expression of NNT-AS1 was associated with increased invasion in CRC. (D) The expression of NNT-AS1 was compared between lymph node metastasis and non-lymph node metastasis. **P<0.01 and ***P<0.001. NNT-AS1, lncRNA nicotinamide nucleotide transhydrogenase antisense RNA 1; CRC, colorectal cancer; N+, lymph node metastasis; N-, non-lymph node metastasis; NC, normal controls.
Expression levels of serum NNT-AS1 in patients with CRC
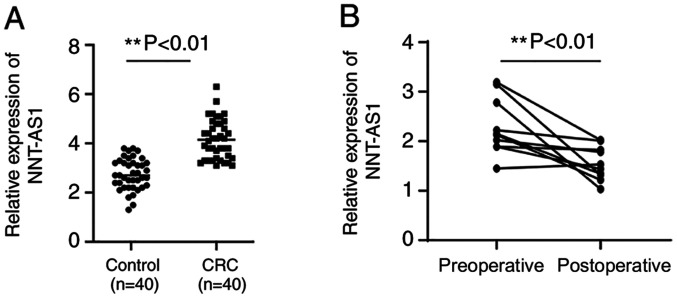
Serum NNT-AS1 expression levels were assayed in patients with CRC and healthy controls. Serum levels of NNT-AS1 were significantly higher in patients compared with healthy controls (P<0.01, Fig. 2A). No significant associations were observed between serum NNT-AS1 levels and clinical features such as gender, age, tumor size and location, local invasion, lymph node metastasis and stage (P>0.05, Table I). The diagnostic value of serum NNT-AS1 levels was then assayed in patients with CRC.
Figure 2.
Expression of NNT-AS1 is higher in the serum of patients with CRC and is decreased after surgery. (A) The expression of NNT-AS1 in serum was assayed in the serum of patients with CRC and healthy normal controls. (B) The expression of NNT-AS1 in serum was compared between preoperative and postoperative samples. **P<0.01. NNT-AS1, lncRNA nicotinamide nucleotide transhydrogenase antisense RNA 1; CRC, colorectal cancer.
Serum NNT-AS1 levels between preoperative and postoperative samples
To examine the changes in NNT-AS1 expression levels after surgery, the expression levels of NNT-AS1 in eight paired preoperative and postoperative serum samples were compared. The results indicated that the expression levels of NNT-AS1 were significantly decreased in postoperative samples compared with preoperative samples (P<0.01, Fig. 2B).
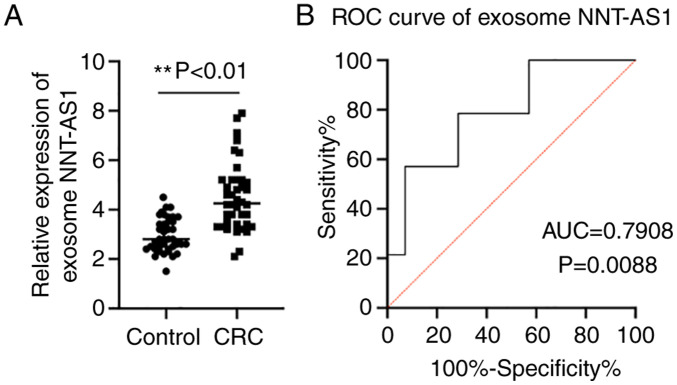
Exosome NNT-AS1 expression levels in patients with CRC and healthy controls
Accumulating evidence has demonstrated that exosomes serve an essential role in maintaining the stability of lncRNAs (13). Thus, the expression levels of NNT-AS1 in exosomes was assayed. The exosome levels of NNT-AS1 in the same cohort of patients with CRC and healthy controls were compared by RT-qPCR. The results demonstrated that NNT-AS1 expression was upregulated in CRC exosomes compared with the controls (P<0.01, Fig. 3A). Then, ROC curve analyses were performed to further elucidate the potential diagnostic value of serum exosomal NNT-AS1. The AUC of serum exosomal NNT-AS1 was 0.7908 (95% CI, 0.6226-0.9590; P=0.0088; Fig. 3B).
Figure 3.
Expression levels of exosomal NNT-AS1 in patients with CRC. (A) The expression levels of exosomal NNT-AS1 were measured in patients with CRC and healthy controls. (B) ROC analysis of exosome NNT-AS1 in patients with CRC. **P<0.01. NNT-AS1, lncRNA nicotinamide nucleotide transhydrogenase antisense RNA 1; CRC, colorectal cancer; ROC, receiver operating characteristic; AUC, area under the curve.
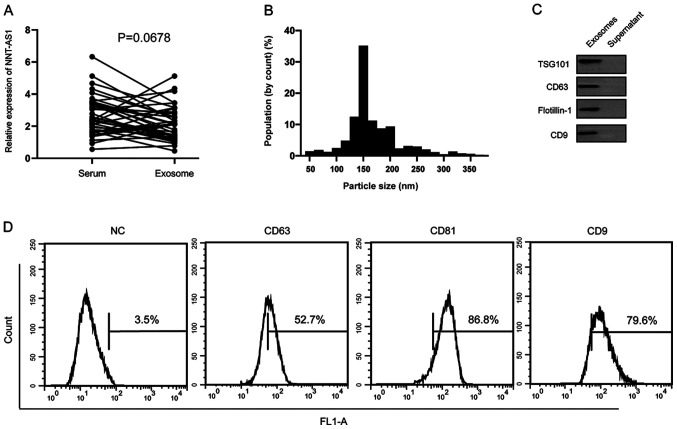
Serum NNT-AS1 is contained in exosomes
It was hypothesized that serum NNT-AS1 is secreted from tumor cells or is protected by exosomes. The levels of NNT-AS1 were measured in 40 pairs of serum samples and exosomes isolated from the same volume of serum. The results demonstrated that there were no significant differences in NNT-AS1 expression levels between serum and exosomes (P=0.0678; Fig. 4A). To further identify the isolated particles, nanoparticle tracking analysis was performed to measure the concentration and size range of the particles. The data demonstrated that the diameters of 10% of the particles were ~100 nm (Fig. 4B). Western blotting and flow cytometry analyses further confirmed the presence of the surface biomarkers of exosomes (Fig. 4C and D).
Figure 4.
Comparison of NNT-AS1 levels between serum samples and exosomes. (A) The levels of NNT-AS1 were compared between serum samples and exosomes and no significant difference was observed. (B) Nanoparticle tracking analysis results demonstrated that the diameters of most particles were within the range of 30–200 nm. (C) Western blotting analysis of exosomal markers. (D) Flow cytometry analysis of surface molecules on exosomes. NNT-AS1, lncRNA nicotinamide nucleotide transhydrogenase antisense RNA 1; TSG101, tumor susceptibility gene 101 protein; NC, normal control.
Knockdown of NNT-AS1 inhibits the proliferation, migration and invasion of CRC cells
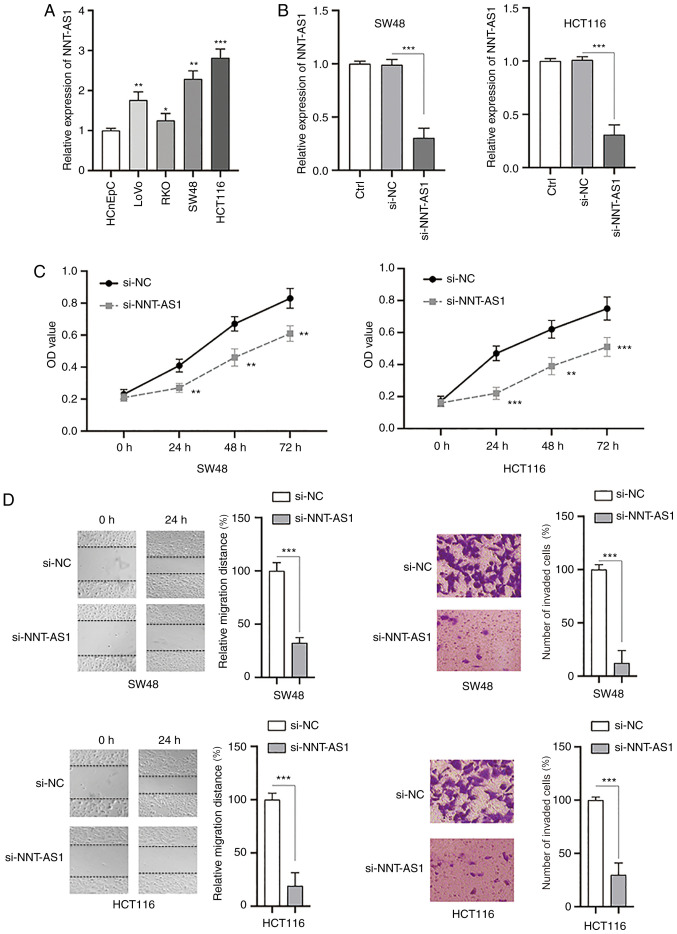
The biological functions of NNT-AS1 in CRC cells were investigated in vitro. The levels of NNT-AS1 in human colorectal normal cells (HCnEpC) and CRC cell lines (LoVo, RKO, SW48 and HCT116) were measured by RT-qPCR. It was identified that the levels of NNT-AS1 were significantly higher in CRC cells compared with normal cells (P<0.05, Fig. 5A). Next, siRNA (si-NNT-AS1) was used to successfully silence the expression of NNT-AS1 in SW48 and HCT116 cells compared with the controls (P<0.001; Fig. 5B). MTT assays demonstrated that the proliferation of both SW48 and HCT116 cells was significantly inhibited after silencing NNT-AS1 (P<0.01; Fig. 5C). The migration and invasion of CRC cells were also significantly decreased after knockdown of NNT-AS1 (P<0.001; Fig. 5D). Taken together, these data suggested that NNT-AS1 may function as an oncogene in CRC cells.
Figure 5.
Inhibition of NNT-AS1 suppresses the proliferation, migration and invasion of CRC cells. (A) The expression of NNT-AS1 was measured in normal colorectal cells (HCnEpC cells) and CRC cells (LoVo, RKO, SW48 and HCT116). (B) CRC cells (SW48 and HCT116) were transfected with the indicated siRNAs and the expression of NNT-AS1 was measured. (C) CRC cells were treated as indicated and cell viability was measured at different time points. (D) CRC cells were treated as indicated and the migration (left) and invasion (right) of cells were investigated. Data are presented as the mean ± standard deviation. *P<0.05, **P<0.01 and ***P<0.001 vs. HCnEpC or si-NC groups or as indicated. All experiments were conducted at least three times. NNT-AS1, lncRNA nicotinamide nucleotide transhydrogenase antisense RNA 1; CRC, colorectal cancer; si-, small interfering RNA; NC, negative control.
NNT-AS1 acts as a sponge of miR-496
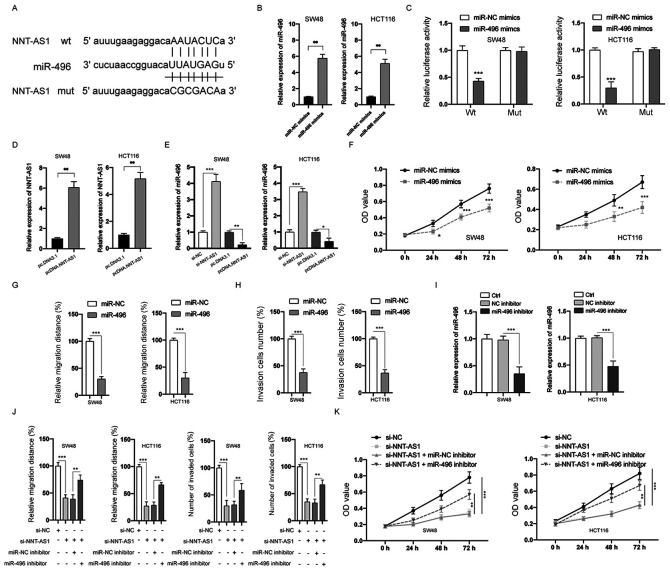
Substantial evidence has indicated that various lncRNAs can act as a sponge to bind and negatively regulate the expression of miRNAs (7). Therefore, bioinformatic tools (StarBase 2.0 and TargetScan) were used to predict the potential targets of NNT-AS1. miR-496 was identified and the putative binding sites between NNT-AS1 and miR-496 are indicated in Fig. 6A. Transfection of miR-496 mimics successfully upregulated the expression of miR-496 in both SW48 and HCT116 cells (P<0.01; Fig. 6B). The interaction between NNT-AS1 and miR-496 was then examined by the luciferase activity assay. The data indicated that miR-496 mimics significantly suppressed the luciferase activity of CRC cells transfected with wt NNT-AS1, whereas those transfected with mut NNT-AS1 were not affected (P>0.05; Fig. 6C). Next, NNT-AS1 was overexpressed by transfection of pcDNA3.NNT-AS1 in SW48 and HCT116 cells (P<0.01; Fig. 6D). In addition, the expression of miR-496 was significantly inhibited by the overexpression of NNT-AS1 and increased by the silencing of NNT-AS1 in SW48 and HCT116 cells (P<0.05; Fig. 6E). MTT assay demonstrated that overexpression of miR-496 inhibited the proliferation of CRC cells, which was similar to the effects of NNT-AS1 downregulation (P<0.05; Fig. 6F). Overexpression of miR-496 also inhibited the migration (P<0.001; Figs. 6G and S1A) and invasion (P<0.001; Figs. 6H and S1B) of CRC cells. To further investigate the function of miR-496 in CRC cells, transfection with an miR-496 inhibitor, which successfully decreased the expression of miR-496, was conducted in CRC cells (P<0.001; Fig. 6I). The effects of silencing NNT-AS1 on the migration, invasion (P<0.01; Figs. 6J and S2A and B) and proliferation (P<0.01; Fig. 6K) of CRC cells could be partially attenuated by inhibition of miR-496. A previous study identified that NNT-AS1 could also bind and inhibit the expression of miR-320 (26). However, no significant difference was observed between the expression of miR-320 in CRC and adjacent normal tissues (data not shown). Taken together, these data suggested that NNT-AS1 functions as a sponge of miR-496.
Figure 6.
NNT-AS1 is a sponge of miR-496. (A) The predicted miR-496 binding sites on wt or mut NNT-AS1. (B) CRC cells were transfected as indicated, and the expression of miR-496 was measured via RT-qPCR. (C) A luciferase reporter assay was performed to measure the luciferase activity in CRC cells after co-transfection of miRNA mimics and NNT-AS1 wt or mut. (D) CRC cells were transfected as indicated, and the expression of NNT-AS1 was measured via RT-qPCR. (E) CRC cells were transfected as indicated and the expression of miR-496 was measured. (F) CRC cells were transfected as indicated and cell viability was measured by the MTT assay. (G) Cell migration was measured. (H) Cell invasion was measured. (I) The levels of miR-496 in CRC were measured. (J) CRC cells were transfected as indicated and the migration and invasion of cells were measured. (K) CRC cells were transfected as indicated and cell viability was measured. Data are presented as the mean ± standard deviation. *P<0.05, **P<0.01 and ***P<0.001 vs. miR-NC mimics or as indicated. All experiments were conducted at least three times. NNT-AS1, lncRNA nicotinamide nucleotide transhydrogenase antisense RNA 1; CRC, colorectal cancer; miR, microRNA; NC, negative control; si-, small interfering RNA; wt, wild-type; mut, mutant; pcD, overexpression vector; RT-qPCR, reverse transcription-quantitative PCR.
RAP2C is a direct target of miR-496
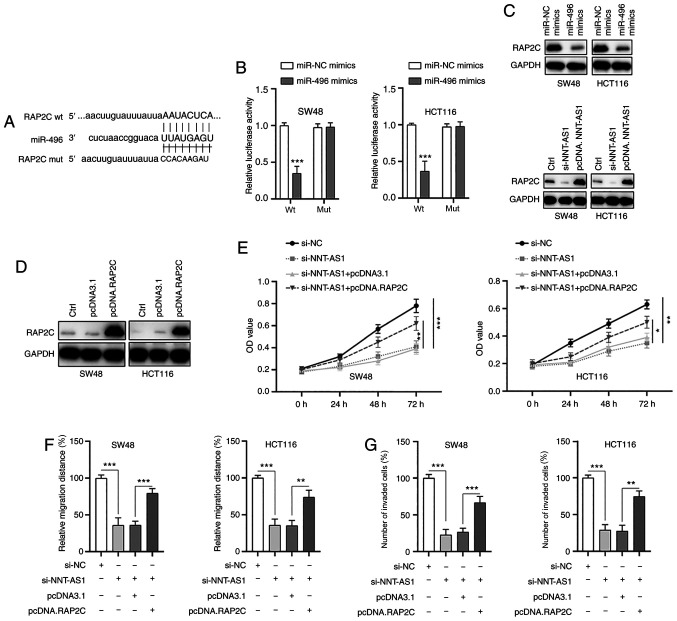
Next, the possible targets of miR-496 were examined. Through bioinformatics analysis, RAP2C was identified as a possible direct target of miR-496 and the putative binding sites are indicated in Fig. 7A. The possible interaction between miR-496 and RAP2C was then validated by the dual-luciferase reporter assay. The results demonstrated that the luciferase activity of wt RAP2C was significantly inhibited by miR-496 mimics, whereas the luciferase activity of mut RAP2C was not affected (Fig. 7B). Furthermore, transfection of miR-496 mimics suppressed the expression of RAP2C (Fig. 7C, top panel) and downregulation or upregulation of NNT-AS1 resulted in the inhibition or increase of RAP2C (Fig. 7C, bottom panel), respectively. These findings suggested that NNT-AS1 may exert its effects via regulation of the miR-496/RAP2C axis. To further examine the function of RAP2C, RAP2C was overexpressed in CRC cells via transfection of a vector containing RAP2C (Fig. 7D). It was identified that the upregulation of RAP2C partially abrogated the effects of NNT-AS1 silencing on the proliferation of CRC cells (Fig. 7E). In addition, overexpression of RAP2C also partially rescued the inhibition of migration (Figs. 7F and S3A) and invasion (Figs. 7G and S3B) of CRC cells caused by NNT-AS1 knockdown. Taken together, these data suggested that RAP2C was a direct target of miR-496 and that NNT-AS1 exerted its effects via regulation of the miR-496/RAP2C axis.
Figure 7.
RAP2C is a direct target of miR-496. (A) The constructed luciferase reporter plasmids containing the predicted wt or mut miR-496 binding sites on RAP2C. (B) A luciferase reporter assay was performed to measure the luciferase activity in CRC cells after co-transfection of miRNA mimics and RAP2C wt or mut. (C) CRC cells were transfected with miR-496 mimics, si-NNT-ASI or pcDNA.NNT-ASI and the levels of the indicated proteins were measured by western blotting. (D) RAP2C was overexpressed in CRC cells by transfection with pcDNA.RAP2C. (E) CRC cells were transfected as indicated and cell viability was measured by an MTT assay. CRC cells were transfected as indicated and (F) migration and (G) invasion were measured. Data are presented as the mean ± standard deviation. *P<0.05, **P<0.01 and ***P<0.001 vs. miR-NC mimics or as indicated. All experiments were conducted at least three times. RAP2C, Ras-related protein Rap-2c; miR, microRNA; wt, wild-type; mut, mutant; NC, negative control; si-, small interfering RNA; pcDNA, overexpression vector; CRC, colorectal cancer; NNT-AS1, lncRNA nicotinamide nucleotide transhydrogenase antisense RNA 1.
Discussion
CRC is one of the most common malignancies worldwide, with considerable mortality rates. A lack of early diagnosis results in limited effective therapeutic options and poor clinical outcomes (3). Therefore, the identification of novel sensitive and specific biomarkers for CRC is urgently needed. Accumulating evidence has indicated that due to different expression patterns in various cancers, lncRNAs can be applied as effective diagnostic and prognostic biomarkers (6–8). In addition, the detection of circulating lncRNAs can be convenient and non-invasive, making lncRNAs an ideal tool for tumor diagnosis. For instance, it was reported that lncRNA AFAP1-AS1 is upregulated in patients with non-small cell lung cancer and demonstrates a higher sensitivity and specificity when compared with healthy controls (27). Wang et al (28) identified that circulating lncRNA CCAT2 is significantly upregulated in the serum of patients with CRC and could be applied as a potential predictor in CRC. The present study focused on NNT-AS1 to study the potential clinical value of circulating NNT-AS1 in CRC.
The lncRNA NNT-AS1, located on chromosome 5p12, has been identified to be differentially expressed in different types of cancers. For example, NNT-AS1 has been identified to contribute to the carcinogenesis of hepatocellular cancer cells through the regulation of miR-363 (29). High expression of NNT-AS1 also leads to the proliferation and invasion of cervical cancer via activation of the Wnt/β-catenin signaling pathway (8). By contrast, Huang et al (30) reported that NNT-AS1 is downregulated in ovarian cancer tissues and cells. These discrepancies indicate the complex role of NNT-AS1 in various cancers and further investigation is necessary to reveal the biological function of NNT-AS1. The present study identified that NNT-AS1 was significantly upregulated in CRC tissues compared with adjacent normal tissues. Notably, the higher expression levels of NNT-AS1 were associated with advanced TNM stage, increased local invasion and positive lymph node metastasis. The findings of the present study are similar to those of a previous study that demonstrated that NNT-AS1 is upregulated in CRC cells and serves an essential role in proliferation, migration and invasion (12). Accumulating evidence has demonstrated that circulating lncRNAs, such as H19, ATB, CCAT1, HOTAIR and GAS5, can be stably detected and used as non-invasive biomarkers for various tumors (31–33). However, to the best of the authors' knowledge, the present study was the first to investigate the levels of NNT-AS1 in the serum of patients with CRC. As a result, it was identified that serum NNT-AS1 levels were also significantly higher in patients with CRC compared with healthy controls. However, no significant association was identified between serum NNT-AS1 levels and other clinicopathological features. The present study also investigated the serum NNT-AS1 expression levels in paired preoperative and postoperative samples and identified that the levels of NNT-AS1 in serum were significantly decreased in the postoperative samples.
A number of studies have reported that lncRNAs stably exist in circulating blood and are protected by exosomes (34,35). Exosomes are type of membrane vesicle with a diameter of 40–150 nm and can be purified by sucrose gradients at a density range of 1.13-1.19 g/ml (35). Exosomes also possess a bilayer lipid membrane that is structurally similar to the cell membrane (36). Exosomes have become a focus of investigation due to their potential as a novel approach for the diagnosis of cancers (37). ncRNAs (including miRNAs and lncRNAs) and proteins from exosomes have been proven to be useful as biomarkers (37). Thus far, only a few lncRNAs have been identified to be contained in exosomes and possess biomarker functions in CRC (28,38). To further confirm the vital function of NNT-AS1 in the diagnosis of CRC, the expression of NNT-AS1 was assayed in exosomes and it was identified that NNT-AS1 was also upregulated in CRC exosomes. The expression of NNT-AS1 in serum samples and in exosomes isolated from the same volume of serum was also compared. No significant differences were observed; therefore, NNT-AS1 might exist in exosomes, which may provide protection to increase the stability of NNT-AS1 in serum.
The functional roles of NNT-AS1 in CRC cells were also examined. The data demonstrated that silencing NNT-AS1 inhibited the proliferation, migration and invasion of CRC cells. These findings are in line with previous studies showing that NNT-AS1 promotes the proliferation and metastasis of various cancers, such as cervical cancer, CRC and liver cancer (8,12,29). Increasing evidence has confirmed that lncRNAs can bind and negatively regulate the function of miRNAs (5–7). The present study revealed that NNT-AS1 acted as a sponge of miR-496, which acted as a tumor suppressor in CRC cells. Previous studies have suggested that miR-496 functions as a tumor suppressor in various cancers, such as CRC, lung cancer and osteosarcoma (18,39,40). A recent study identified that miR-496 acted as an oncogene and promoted malignancy in CRC cells (41). This discrepancy might be due to different cancer types and further investigation is needed to further verify the role of miR-496 in more cancer types. The present study also revealed that RAP2C might be a direct target of miR-496. RAP2C, a member of the Ras family, has been identified to participate in the tumorigenesis of various cancers. For instance, RAP2C acts as an oncogene in human osteosarcoma cells (20). Inhibition of RAP2C suppresses the proliferation and promotes the apoptosis of breast cancer cells (21). However, there is little information concerning the role of RAP2C in CRC. Overexpression of RAP2C partially abrogated the effects of NNT-AS1 knockdown and rescued the proliferation, migration and invasion of CRC cells. The data from the present study suggested that RAP2C acted as an oncogene in CRC cells and further investigation is needed to confirm this.
In summary, the data from the present study suggested that NNT-AS1 expression is significantly upregulated in tissue and serum samples of patients with CRC. Circulating NNT-AS1 may be protected by exosomes and could be used as a potential predictor in CRC. It was also identified that NNT-AS1 regulated the proliferation, migration and invasion of CRC cells via the miR-496/RAP2C axis. The results from the present study identified novel pathways regulating CRC progression, which may facilitate the identification of novel therapeutic targets for CRC.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by Medical Scientific Research Identification of Zhejiang Province, China (grant. no. 2019KY595), Provincial Natural Fund of Zhejiang Province (grant. no. LGC20H160002) and Ningbo Clinical Research Center for Digestive System Tumors (grant. no. 2019A21003).
Funding
The present study was supported by Medical Scientific Research Identification of Zhejiang Province, China (grant. no. 2019KY595), Provincial Natural Fund of Zhejiang Province (grant. no. LGC20H160002) and Ningbo Clinical Research Center for Digestive System Tumors (grant. no. 2019A21003).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HY and JH performed the experiments. ZY repeated the experiments (data not shown), validated the data and conducted the statistical analysis. SC analyzed and interpreted the data. ZY and SC confirm the authenticity of all the raw data. YC conceived and designed the study, and drafted and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethical Committee of Hwamei Hospital, University of Chinese Academy of Sciences (Ningbo, China) and all procedures were conducted according to the Declaration of Helsinki. Written informed consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 2.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 3.The Lancet. Toward better control of colorectal cancer. Lancet. 2014;383:1437. doi: 10.1016/S0140-6736(14)60699-1. [DOI] [PubMed] [Google Scholar]
- 4.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667. doi: 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han D, Wang M, Ma N, Xu Y, Jiang Y, Gao X. Long noncoding RNAs: Novel players in colorectal cancer. Cancer Lett. 2015;361:13–21. doi: 10.1016/j.canlet.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hua F, Liu S, Zhu L, Ma N, Jiang S, Yang J. Highly expressed long non-coding RNA NNT-AS1 promotes cell proliferation and invasion through Wnt/β-catenin signaling pathway in cervical cancer. Biomed Pharmacother. 2017;92:1128–1134. doi: 10.1016/j.biopha.2017.03.057. [DOI] [PubMed] [Google Scholar]
- 9.Huang Q, Wang S, Li X, Yang F, Feng C, Zhong K, Qiu M, Wang J. Circular RNA ATXN7 is upregulated in non-small cell lung cancer and promotes disease progression. Oncol Lett. 2019;17:4803–4810. doi: 10.3892/ol.2019.10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Ma H, Kong W, Liu B, Zhang X. Up-regulated circular RNA VANGL1 contributes to progression of non-small cell lung cancer through inhibition of miR-195 and activation of Bcl-2. Biosci Rep. 2019;39:BSR20182433. doi: 10.1042/BSR20182433. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.He W, Zhang Y, Xia S. LncRNA NNT-AS1 promotes non-small cell lung cancer progression through regulating miR-22-3p/YAP1 axis. Thorac Cancer. 2020;11:549–560. doi: 10.1111/1759-7714.13280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Q, Yang L, Hu X, Jiang Y, Hu Y, Liu Z, Liu J, Wen T, Ma Y, An G, Feng G. Upregulated NNT-AS1, a long noncoding RNA, contributes to proliferation and migration of colorectal cancer cells in vitro and in vivo. Oncotarget. 2017;8:3441–3453. doi: 10.18632/oncotarget.13840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun T, Kalionis B, Lv G, Xia S, Gao W. Role of exosomal noncoding RNAs in lung carcinogenesis. Biomed Res Int. 2015;2015:125807. doi: 10.1155/2015/125807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7:e30679. doi: 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luga V, Zhang L, Viloria-Petit AM, Ogunjimi AA, Inanlou MR, Chiu E, Buchanan M, Hosein AN, Basik M, Wrana JL. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell. 2012;151:1542–1556. doi: 10.1016/j.cell.2012.11.024. [DOI] [PubMed] [Google Scholar]
- 16.Hewson C, Morris KV. Form and function of exosome-associated long non-coding RNAs in cancer. Curr Top Microbiol Immunol. 2016;394:41–56. doi: 10.1007/82_2015_486. [DOI] [PubMed] [Google Scholar]
- 17.Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma R, Zhu P, Liu S, Gao B, Wang W. MiR-496 suppress tumorigenesis via targeting BDNF-mediated PI3K/Akt signaling pathway in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;518:273–277. doi: 10.1016/j.bbrc.2019.08.046. [DOI] [PubMed] [Google Scholar]
- 19.Alvarado S, Wyglinski J, Suderman M, Andrews SA, Szyf M. Methylated DNA binding domain protein 2 (MBD2) coordinately silences gene expression through activation of the microRNA hsa-mir-496 promoter in breast cancer cell line. PLoS One. 2013;8:e74009. doi: 10.1371/journal.pone.0074009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu J, Du W, Wang X, Wei L, Pan Y, Wu X, Zhang J, Pei D. Ras-related protein Rap2c promotes the migration and invasion of human osteosarcoma cells. Oncol Lett. 2018;15:5352–5358. doi: 10.3892/ol.2018.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu X, Qiu J, Zhang T, Yang Y, Guo S, Li T, Jiang K, Zahoor A, Deng G, Qiu C. MicroRNA-188-5p promotes apoptosis and inhibits cell proliferation of breast cancer cells via the MAPK signaling pathway by targeting Rap2c. J Cell Physiol. 2020;235:2389–2402. doi: 10.1002/jcp.29144. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, Huang C, Zhang A, Lu C, Liu L. Overexpression of circRNA_100290 promotes the progression of laryngeal squamous cell carcinoma through the miR-136-5p/RAP2C axis. Biomed Pharmacother. 2020;125:109874. doi: 10.1016/j.biopha.2020.109874. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Yu R, Yu BX, Chen JF, Lv XY, Yan ZJ, Cheng Y, Ma Q. Anti-tumor effects of Atractylenolide I on bladder cancer cells. J Exp Clin Cancer Res. 2016;35:40. doi: 10.1186/s13046-016-0312-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muschelli J. ROC and AUC with a binary predictor: Potentially a misleading metric. J Classif. 2002;37:696–708. doi: 10.1007/s00357-019-09345-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li C, Zhang S, Qiu T, Wang Y, Ricketts DM, Qi C. Upregulation of long non-coding RNA NNT-AS1 promotes osteosarcoma progression by inhibiting the tumor suppressive miR-320a. Cancer Biol Ther. 2019;20:413–422. doi: 10.1080/15384047.2018.1538612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li T, Ding ZL, Zheng YL, Wang W. MiR-484 promotes non-small-cell lung cancer (NSCLC) progression through inhibiting Apaf-1 associated with the suppression of apoptosis. Biomed Pharmacother. 2017;96:153–164. doi: 10.1016/j.biopha.2017.09.102. [DOI] [PubMed] [Google Scholar]
- 28.Wang L, Duan W, Yan S, Xie Y, Wang C. Circulating long non-coding RNA colon cancer-associated transcript 2 protected by exosome as a potential biomarker for colorectal cancer. Biomed Pharmacother. 2019;113:108758. doi: 10.1016/j.biopha.2019.108758. [DOI] [PubMed] [Google Scholar]
- 29.Lu YB, Jiang Q, Yang MY, Zhou JX, Zhang Q. Long noncoding RNA NNT-AS1 promotes hepatocellular carcinoma progression and metastasis through miR-363/CDK6 axis. Oncotarget. 2017;8:88804–88814. doi: 10.18632/oncotarget.21321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Y, Shi J, Xu Y. Long non-coding RNA NNT-AS1 contributes to cell proliferation, metastasis and apoptosis in human ovarian cancer. Oncol Lett. 2018;15:9264–9270. doi: 10.3892/ol.2018.8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abedini P, Fattahi A, Agah S, Talebi A, Beygi AH, Amini SM, Mirzaei A, Akbari A. Expression analysis of circulating plasma long noncoding RNAs in colorectal cancer: The relevance of lncRNAs ATB and CCAT1 as potential clinical hallmarks. J Cell Physiol. 2019;234:22028–22033. doi: 10.1002/jcp.28765. [DOI] [PubMed] [Google Scholar]
- 32.Zhang R, Xia Y, Wang Z, Zheng J, Chen Y, Li X, Wang Y, Ming H. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem Biophys Res Commun. 2017;490:406–414. doi: 10.1016/j.bbrc.2017.06.055. [DOI] [PubMed] [Google Scholar]
- 33.Shen J, Hodges TR, Song R, Gong Y, Calin GA, Heimberger AB, Zhao H. Serum HOTAIR and GAS5 levels as predictors of survival in patients with glioblastoma. Mol Carcinog. 2018;57:137–141. doi: 10.1002/mc.22739. [DOI] [PubMed] [Google Scholar]
- 34.Arita T, Ichikawa D, Konishi H, Komatsu S, Shiozaki A, Shoda K, Kawaguchi T, Hirajima S, Nagata H, Kubota T, et al. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013;33:3185–3193. [PubMed] [Google Scholar]
- 35.Niu L, Song X, Wang N, Xue L, Song X, Xie L. Tumor-derived exosomal proteins as diagnostic biomarkers in non-small cell lung cancer. Cancer Sci. 2019;110:433–442. doi: 10.1111/cas.13862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pant S, Hilton H, Burczynski ME. The multifaceted exosome: Biogenesis, role in normal and aberrant cellular function and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol. 2012;83:1484–1494. doi: 10.1016/j.bcp.2011.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou R, Chen KK, Zhang J, Xiao B, Huang Z, Ju C, Sun J, Zhang F, Lv XB, Huang G. The decade of exosomal long RNA species: An emerging cancer antagonist. Mol Cancer. 2018;17:75. doi: 10.1186/s12943-018-0823-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu T, Zhang X, Gao S, Jing F, Yang Y, Du L, Zheng G, Li P, Li C, Wang C. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget. 2016;7:85551–85563. doi: 10.18632/oncotarget.13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi NN, Tian S, Li X, Wang FL, Liu B. Up-regulation of microRNA-496 suppresses proliferation, invasion, migration and in vivo tumorigenicity of human osteosarcoma cells by targeting eIF4E. Biochimie. 2019;163:1–11. doi: 10.1016/j.biochi.2019.04.017. [DOI] [PubMed] [Google Scholar]
- 40.Ye J, Xie W, Zuo Y, Jing G, Tong J. MicroRNA-496 suppresses tumor cell proliferation by targeting BDNF in osteosarcoma. Exp Ther Med. 2020;19:1425–1431. doi: 10.3892/etm.2019.8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, Yan B, Zhang P, Liu S, Li Q, Yang J, Yang F, Chen E. MiR-496 promotes migration and epithelial-mesenchymal transition by targeting RASSF6 in colorectal cancer. J Cell Physiol. 2020;235:1469–1479. doi: 10.1002/jcp.29066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.