Abstract
Long non-coding RNAs (lncRNAs) regulate gene expression in a variety of ways at epigenetic, chromatin remodeling, transcriptional, and translational levels. Accumulating evidence suggests that lncRNA X-inactive specific transcript (lncRNA Xist) serves as an important regulator of cell growth and development. Despites its original roles in X-chromosome dosage compensation, lncRNA Xist also participates in the development of tumor and other human diseases by functioning as a competing endogenous RNA (ceRNA). In this review, we comprehensively summarized recent progress in understanding the cellular functions of lncRNA Xist in mammalian cells and discussed current knowledge regarding the ceRNA network of lncRNA Xist in various diseases. Long non-coding RNAs (lncRNAs) are transcripts that are more than 200 nt in length and without an apparent protein-coding capacity (Furlan and Rougeulle, 2016; Maduro et al., 2016). These RNAs are believed to be transcribed by the approximately 98–99% non-coding regions of the human genome (Derrien et al., 2012; Fu, 2014; Montalbano et al., 2017; Slack and Chinnaiyan, 2019), as well as a large variety of genomic regions, such as exonic, tronic, and intergenic regions. Hence, lncRNAs are also divided into eight categories: Intergenic lncRNAs, Intronic lncRNAs, Enhancer lncRNAs, Promoter lncRNAs, Natural antisense/sense lncRNAs, Small nucleolar RNA-ended lncRNAs (sno-lncRNAs), Bidirectional lncRNAs, and non-poly(A) lncRNAs (Ma et al., 2013; Devaux et al., 2015; St Laurent et al., 2015; Chen, 2016; Quinn and Chang, 2016; Richard and Eichhorn, 2018; Connerty et al., 2020). A range of evidence has suggested that lncRNAs function as key regulators in crucial cellular functions, including proliferation, differentiation, apoptosis, migration, and invasion, by regulating the expression level of target genes via epigenomic, transcriptional, or post-transcriptional approaches (Cao et al., 2018). Moreover, lncRNAs detected in body fluids were also believed to serve as potential biomarkers for the diagnosis, prognosis, and monitoring of disease progression, and act as novel and potential drug targets for therapeutic exploitation in human disease (Jiang W. et al., 2018; Zhou et al., 2019a). Long non-coding RNA X-inactive specific transcript (lncRNA Xist) are a set of 15,000–20,000 nt sequences localized in the X chromosome inactivation center (XIC) of chromosome Xq13.2 (Brown et al., 1992; Debrand et al., 1998; Kay, 1998; Lee et al., 2013; da Rocha and Heard, 2017; Yang Z. et al., 2018; Brockdorff, 2019). Previous studies have indicated that lncRNA Xist regulate X chromosome inactivation (XCI), resulting in the inheritable silencing of one of the X-chromosomes during female cell development. Also, it serves a vital regulatory function in the whole spectrum of human disease (notably cancer) and can be used as a novel diagnostic and prognostic biomarker and as a potential therapeutic target for human disease in the clinic (Liu et al., 2018b; Deng et al., 2019; Dinescu et al., 2019; Mutzel and Schulz, 2020; Patrat et al., 2020; Wang et al., 2020a). In particular, lncRNA Xist have been demonstrated to be involved in the development of multiple types of tumors including brain tumor, Leukemia, lung cancer, breast cancer, and liver cancer, with the prominent examples outlined in Table 1. It was also believed that lncRNA Xist (Chaligne and Heard, 2014; Yang Z. et al., 2018) contributed to other diseases, such as pulmonary fibrosis, inflammation, neuropathic pain, cardiomyocyte hypertrophy, and osteoarthritis chondrocytes, and more specific details can be found in Table 2. This review summarizes the current knowledge on the regulatory mechanisms of lncRNA Xist on both chromosome dosage compensation and pathogenesis (especially cancer) processes, with a focus on the regulatory network of lncRNA Xist in human disease.
Keywords: long non-coding RNA, lncRNA Xist, cancer, disease, X-chromosome inactivation, X-chromosome inactivation center
The Role of LncRNA Xist in X Chromosome Dosage Compensation
In most mammals, sex is determined by a system based on X and Y chromosomes (Deng et al., 2014), with males holding the XY chromosome and females XX. Dosage compensation is thus needed to ensure equivalent expression levels of sex-linked and autosomal genes (Polito et al., 1990; Bone and Kuroda, 1996; Larsson and Meller, 2006; Disteche, 2012, 2016; Ferrari et al., 2014) despite the presence of an extra X-chromosome in female cells (Deng et al., 2014). X-chromosome inactivation (XCI), which refers to the random selection and transcriptional silence of one of two X-chromosomes in females at the early stages of embryonic development, is a unique dosage compensation mechanism in mammals (Waldron, 2016; Bar et al., 2019; Strehle and Guttman, 2020; Yu B. et al., 2020). In most placental mammals, there are two waves of XCI: the imprinted XCI exists in the fertilized embryo and extraembryonic tissues, and the random XCI persists in the inner cell mass (after implantation around embryonic day 5.5), yet humans lack the imprinted XCI and instead have X chromosome dampening (XCD) (Ropers et al., 1978; Harper et al., 1982; Kung et al., 2013; Lee et al., 2013; van Bemmel et al., 2016; Finestra and Gribnau, 2017; Sahakyan et al., 2018).
XCI is subdivided into distinct phases: initiation, establishment, and maintenance of the inactive X-chromosome (Gontan et al., 2011; Maduro et al., 2016). Initiation phase is a stochastic process (Spatz et al., 2004; Maduro et al., 2016; Jegu et al., 2017) that involves X-X pairing, counting, and XCI activation (xist activation, etc.) processes, and ensures that any number of X chromosomes randomly generate only one active X chromosome (Xa) expressed in each female cell and inactive X chromosome (Xi) is hetero-chromatinized and silenced in female cells. Establishment phase (Spatz et al., 2004; Maduro et al., 2016; Colognori et al., 2020) involves building a chromosomal memory that persists through the ensuring maintenance phase and ensures stable retention of repressive heterochromatin. Once the establishment phase is completed, the XCI is remarkably stable and becomes more difficult to reactivate. Maintenance phase is keeping the silenced state of XCI after the establishment phase via continuing lncRNA Xist expression. Once Xi is established, the Xi fully maintains its silent configuration and is clonally propagated throughout cell divisions (Maduro et al., 2016; Finestra and Gribnau, 2017). Numorous studies suggest that all three phases of XCI are governed by the lncRNA Xist (Lu et al., 2017; Sahakyan et al., 2018; Sidorenko et al., 2019).
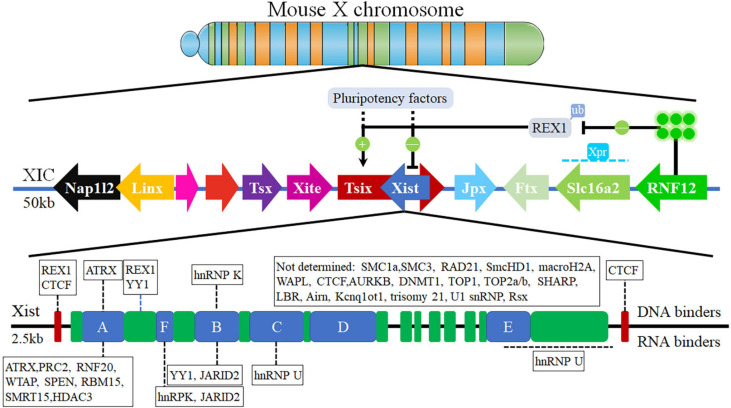
XIC is the X-linked minimal genetic region which contains various factors and genes, such as Xist and Tsix, that are necessary and sufficient to initiate the XCI process in female cells (Willard, 1996; Sherstyuk et al., 2013; Hwang et al., 2015; Loda and Heard, 2019). XIC (Figure 1) is located in 100–500 kb region of mouse X chromosome and 2.3 Mb syntenic region of human X chromosome, and includes a cluster of lncRNA loci, such as Ftx, Jpx, Xist, Tsix, Xite, RepA, and so on (Spatz et al., 2004; Augui et al., 2011; Maduro et al., 2016; Jegu et al., 2017; Loda and Heard, 2019; Sidorenko et al., 2019). lncRNA Xist exists inside XIC, specifically at a location 15 kb downstream from Tsix antisense (Sado and Brockdorff, 2013; Gendrel and Heard, 2014; Mira-Bontenbal and Gribnau, 2016; da Rocha and Heard, 2017; Pintacuda et al., 2017b; Monfort and Wutz, 2020), and contains several functional domains that are a series of conservation repetitive motifs of A-to-F repeats (Figure 1). lncRNA Xist is transcriptionally activated with the initiation of the XIC process and is also believed to contribute to the complete process of XCI as a master regulator.
FIGURE 1.
The X-chromosome Inactivation Center (Maduro et al., 2016). The X inactivation center consists of the different genes located and multiple genes encoding lncRNA, containing Xist, Tsix, Tsx, Xite, Jpx, Ftx, DNA binders, and RNA binders.
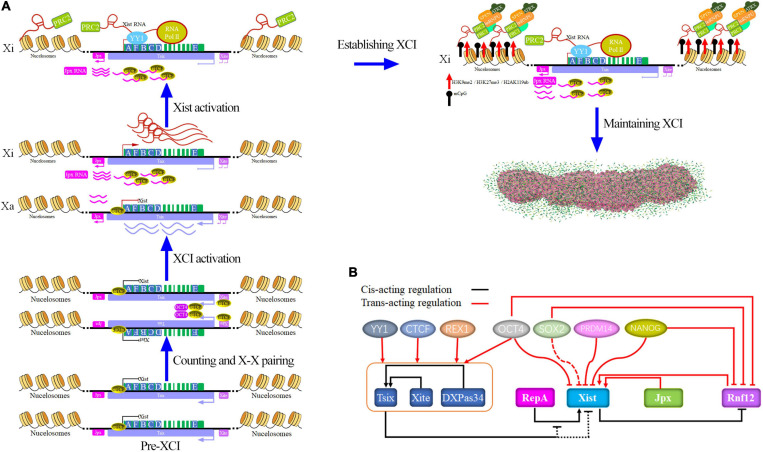
lncRNA Xist and its associated chromatin modifying complex plays a vital role in the regulation of the XCI process (Figure 2). A detailed description of the XCI process is beyond the scope of this review, and more specific detail is given in references (Spatz et al., 2004; Augui et al., 2011; Jegu et al., 2017), We briefly described regulatory process involved in LncRNA Xist in the review (Figure 2). During the initiation phase, the complex factors (OCT4, CTCF, Tsix, Xite, etc.), which separately bind the Xa and Xi, facilitates the X chromosome pairing and counting in the embryo after fertilization (Xu et al., 2006; Donohoe et al., 2009; Kung et al., 2015). After counting and pairing, XCI initiation is also accompanied by Tsix, Xist, etc. upregulation which is controlled by the network of genetic interactions (Figure 2B), such as Tsix, Sox2, PRDM14, OCT4, Jpx, Rnf12, and RepA (Augui et al., 2011; Khamlichi and Feil, 2018). When complete onset of XCI occurs, they employ divergent transcription fates with one becoming the Xa chromosome and the other becoming the Xi chromosome (Jegu et al., 2017). In Xi, lncRNA Xist activation and expression is modulated by numerous factors, such as pluripotency factor (NANOG, OCT4, SOX2, PRDM14, and REX1), RNF12, Tsix, and RepA (PcG protein recruitment), and more information is given in reference (Augui et al., 2011; Khamlichi and Feil, 2018). The regulation of Tsix expression is beyond the scope of this review, and more specific details can be found in references (Willard, 1996; Gontan et al., 2012; Gayen et al., 2015). Once Xist expression has been activated, Xist binds Polycomb repressive complex 2 (PRC2) via Repeat A formed Xist-PRC2 complex, and YY1 tethers the PRC2-Xist complex through Repeat C to the Xi nucleation center which obtains lncRNA Xist-PRC2 complex by the RNA polymerase II (RNA Pol II) (Jeon and Lee, 2011; Thorvaldsen et al., 2011; Makhlouf et al., 2014; Chigi et al., 2017).
FIGURE 2.
Model for Xist and Xist regulation at the process of XCI. (A) The process of dynamic and multifaceted modulation of XCI by lncRNA Xist. lncRNA Xist is a multitasking RNA that recruits protein complexes (such as OCT4, CTCF, Tsix, Xite, PRC1, PRC2, SPEN, ATRX, hnRNPU, hnRNPK, SHARP, HDAC3, LBR, Airn, Kcnq1ot1, RBM15, WTAP, trisomy 21, U1 snRNP, Rsx, Sox2, PRDM14, Jpx, Rnf12, and RepA) to initiate, establish, and maintain the XCI state by histone modifications, DNA methylation, and H4 hypoacetylation. (B) LncRNA Xist regulation network of genetic interactions (Augui et al., 2011). Note that here arrows do not necessarily imply direct regulation.
After the initiation phase, LncRNA Xist recruits protein complex factors excluding RNA Pol II, and induces a global suppression of lncRNA Xist topologically associated domains (TAD), which is involved in epigenetic modification and chromatin compaction to the Xi chromosome to spreads along the Xi at the established phase (Giorgetti et al., 2016; Mira-Bontenbal and Gribnau, 2016; Finestra and Gribnau, 2017; van Bemmel et al., 2019; Galupa et al., 2020). These protein complexes (Figure 2A) include the heterogeneous nuclear protein U (hnRNPU; also known as SAF-A), which is required for lncRNA Xist localization (Hasegawa et al., 2010; Kolpa et al., 2016; Sakaguchi et al., 2016; Loda and Heard, 2019), heterogeneous nuclear ribonucleoprotein K (hnRNPK), which is required for Xist-mediated chromatin modifications and Polycomb recruitment but not lncRNA Xist localization (Chu et al., 2015; Pintacuda et al., 2017a; Loda and Heard, 2019; Wang et al., 2019g), and the gene-silencing factor Spen, which is not required for Xist RNA localization (Chu et al., 2015; Monfort et al., 2015; Loda and Heard, 2019; Dossin et al., 2020) and binds to C, B, F, and A repeats at the 5′ end of the lncRNA Xist. ATRX directs binding to two major Polycomb repressive complexes (PRCs). -PRC1 and -PRC2 are involved in epigenetic silencing (acetylation of histone H3 and H4 and CpG island methylation, etc.) (Sarma et al., 2014; Minajigi et al., 2015; Pinheiro and Heard, 2017; Colognori et al., 2019; Lee et al., 2019; Wang et al., 2019a; Chen and Zhang, 2020). Other protein complexes (Mira-Bontenbal and Gribnau, 2016; Loda and Heard, 2019) also take part in the lncRNA Xist spreading procession, such as SHARP (McHugh et al., 2015), HDAC3 (Zylicz et al., 2019), LBR (Chen C.K. et al., 2016; Nesterova et al., 2019), Airn and Kcnq1ot1 (Schertzer et al., 2019), RBM15 and WTAP (Mira-Bontenbal and Gribnau, 2016), trisomy 21 (Jiang et al., 2013), U1 snRNP (Yin et al., 2020), Rsx (Grant et al., 2012), and CdK8 (Postlmayr et al., 2020). LncRNA Xist recruits repressive complexes, which leads to immediate histone modifications and DNA methylation (such as H2AK119Ub, H3K27me3, and CpG island) and coats on the Xi to build Xi (Mira-Bontenbal and Gribnau, 2016; Pinheiro and Heard, 2017; Wang et al., 2020a). Taken together, the Xi has been established and maintained in an inactive state by continuous synthesis of lncRNA Xist RNA.
The Role of LncRNA Xist in Cancer
Cancer, of which there are over 200 different types, is a complex disease in which cells in a specific tissue are no longer fully responsive to the signals within the tissue that regulate cellular differentiation, survival, proliferation, and death. As a result, these cells accumulate within the tissue, causing local damage and inflammation Cancer cells proliferate (growth) out of control, spread to other tissues (metastasize), and lose the ability to die via the normal process of cell apoptosis (death). The discovery of lncRNA Xist has contributed to cancer development and progression by regulation of the downstream signaling processes (Table 1). This also provides a window into the understanding of aberrant expression of lncRNA Xist associated with tumorigenesis, metastasis, and tumor stage. lncRNA Xist is a novel potential biomarker and potentially could be used in diagnosis and therapy for different types of cancer.
TABLE 1.
LncRNA Xist and miRNA in cancer.
| Cancer type | miRNA | Target | Mechanism of action and function | Signaling pathway | References |
| Bladder cancer | miR-124, miR-139-5p, miR-200c, miR-133a, miR-335 | AR, Wnt1, TET1, p53 | Aberrant expression lncRNA Xist is involved in cancer cells growth, proliferation, metastasis, migration, invasion, apoptosis, epithelial mesenchymal transition and drug resistance | TGF-beta signaling pathway, PIK3/AKT signaling pathway, mTOR signaling pathway, Wnt/ β-catenin signaling pathway, p53 signaling pathway, MAPK signaling pathway, FOXO signaling pathway, HIF-1 signaling pathway, Thyroid hormone signaling pathway, Notch signaling pathway, C-type lectin receptor signaling pathway, JAK-STAT signaling pathway, AGE-RAGE signaling pathway, Pathways of neurodegeneration—multiple diseases and ECM-receptor interaction, etc. | Hu et al., 2017; Xiong et al., 2017; Xu R. et al., 2018; Hu B. et al., 2019; Zhou et al., 2019c; Chen D. et al., 2020 |
| Breast cancer | miR-155, miR-20a, miR-200c-3p, miR-454, miR-92b, miR-503, miR-125b-5p, miR-362-5p | CDX1, TP53, ANLN, Slug, ESA, PHLPP1, AKT, MSN, cMet, NLRC5, UBAP1 | Xing et al., 2018; Zhao L. et al., 2018; Zheng R. et al., 2018; Li et al., 2020c,d,e; Liu et al., 2020a; Zhang et al., 2020a | ||
| Colorectal cancer | miR-137, miR-132-3p, miR-486-5p, miR-93-5p, miR-124, miR-30a-5p, miR-338-3p | EZH2, MAPK1, NRP-2, HIF1A, AXL, METTL14, SGK1, ROR1, PAX5 | Chen D.L. et al., 2017; Song et al., 2017; Liu et al., 2018a, 2019a; Zhu J. et al., 2018; Zhang et al., 2019d; Ma et al., 2020; Yang L.G. et al., 2020 | ||
| Glioblastoma | miR-152, miR-27a, miR-429, miR-137, miR-126, miR-133a, miR-29c, miR-204-5p | Smurf1, ZO-2, FOXC1, Rac1, SLC1A5, IRS1, SOX4, MMR, Bcl-2, ASCT2 | Du P. et al., 2017; Wang Z. et al., 2017; Yu H. et al., 2017; Cheng Z.H. et al., 2020; Luo C.X. et al., 2020; Shen J. et al., 2020; Sun Y. et al., 2020; Yao et al., 2020; Zhao Q. et al., 2020 | ||
| Hepatocellular carcinoma | miR-29b, miR-92b, miR-155-5p, miR-200b-3p, miR-139-5p, miR-194-5p, miR-497-5p, miR-181a | HMGB1, SMAD7, SOx6, PTEN, PDK1, AKT, MAPK1, PDCD4, PTEN | Zhuang et al., 2016; Chang et al., 2017; Mo et al., 2017; Kong et al., 2018; Lin et al., 2018; Liu and Xu, 2019; Xie et al., 2019; Zhang et al., 2019h | ||
| Nasopharyngeal | miR-34a-5p, miR-29c, miR-491-5p, miR-148a-3p, miR-381-3p | E2F3, Notch3, ADAM17, NEK5, PDCD4, Fas-L | Song et al., 2016; Han et al., 2017; Cheng Q. et al., 2018; Shi et al., 2020; Zhao C.H. et al., 2020 | ||
| Lung cancer | miR-140, miR-363-3p, let-7i, miR-449a, miR-374a, miR-212-3p, miR-186-5p, miR-137, miR-744, miR-367, miR-141, miR-16, miR-335, miR-144-3p, miR-17, miR-142-5p | iASPP, TCF-4, MDM2, BAG-1, HIF1A-AS1, KLF2, Bcl-2, LARP1, CBLL1, PXN, Notch-1, RING1, ZEB2, CDK8, SOD2, ROS, SMAD2, p53, NLRP3, MDR1, MRP1, ATG7, PAX6 | Sun J. et al., 2017; Sun W. et al., 2017; Wang H.Y. et al., 2017; Xu Z.Z. et al., 2017; Zhang Y.L. et al., 2017; Jiang H.J. et al., 2018; Li C. et al., 2018; Wang et al., 2018c, 2019c; Liu et al., 2019b; Qiu et al., 2019; Tian et al., 2019; Zhou et al., 2019e; Jiang et al., 2020; Rong et al., 2020; Xu et al., 2020 | ||
| Osteosarcoma | miR-21-5p, miR-193-3p, miR-195-5p, miR-137, miR-302b, miR-375-3p, miR-153 | p21, NF-kB, PUMA, PDCD4, RSf1, YAP, RAP2B, AKT, mTOR, SNAl1 | Wu D.P. et al., 2017; Zhang and Xia, 2017; Lv et al., 2018; Yang C. et al., 2018; Li H. et al., 2019; Sun X. et al., 2019; Wen et al., 2020 | ||
| Pancreatic cancer | miR-133a, miR-140, miR-124, miR-34a-5p, miR-34a, miR-141-3p, miR-429 | EGFR, iASPP, YAP, EGFR, TGF-β2, ZEB1 | Liang et al., 2017; Wei W. et al., 2017; Sun Z. et al., 2018; Shen et al., 2019; Sun and Zhang, 2019; Zou et al., 2020 | ||
| Retinoblastoma | miR-21-5p, miR-124, miR-101, miR-140-5p, miR-200a-3p | VEGF, NKILA, STAT3, ZEB1, ZEB2, SOX4, NRP1 | Hu et al., 2018; Cheng Y. et al., 2019; Dong et al., 2020; Wang et al., 2020c; Zhao H. et al., 2020 | ||
| Cervical cancer | miR-200a, miR-140-5p, miR-889-3p | Fus, ORC1, SIX1 | Zhu H. et al., 2018; Chen X. et al., 2019c; Liu et al., 2020d | ||
| Gastric cancer | miR-101, miR-497, miR-185, miR-337 | EZH2, MACC1, TGF-β1, MDR1, MRP1, JAK2 | Chen D.L. et al., 2016; Ma L. et al., 2017; Zhang Q. et al., 2018; Zheng W. et al., 2020 | ||
| Melanoma | miR-21, miR-139-5p, miR-217 | PI3KR1, ROCK1 | Pan et al., 2019; Zhang et al., 2019f; Tian K. et al., 2020 | ||
| Esophageal cancer | miR-101, miR-494 | EZH2, CDK6 | Wu X. et al., 2017; Chen et al., 2019f | ||
| Laryngeal squamous cell carcinoma | miR-124-3p, miR-144, miR-125-5p | EZH2, IRS1, TRIB2 | Xiao D. et al., 2019; Cui et al., 2020; Liu et al., 2020b | ||
| Ovarian cancer | miR-214-3p, miR-150-5p | PTEN, PDCD4 | Zuo et al., 2019; Wang and Li, 2020 | ||
| Neuroendocrine tumor | miR-424-5p | bFGF | Zhou et al., 2019b | ||
| Neuroblastoma | miR-375 | EZH2, DKK1, L1CAM | Yang H. et al., 2020 | ||
| Thyroid cancer | miR-34a, miR-141 | MET | Liu et al., 2018c; Xu Y. et al., 2018 | ||
| Colon cancer | miR-34a | Wnt/β-catenin | Sun N.N. et al., 2018 | ||
| Renal cell carcinoma | miR-106b-5p, miR-302c | p21, SDC1 | Zhang J. et al., 2017; Sun K. et al., 2019 | ||
| Prostate cancer | miR-23a | RKIP, LINE-1 | Laner et al., 2005; Du Y. et al., 2017 | ||
| Chordoma | miR-124-3p | iASPP | Hai, 2020 |
LncRNA Xist in Bladder Cancer
Bladder cancer is more common in men than in women, with respective incidence and mortality rates of 9.6 and 3.2 per 100,000 in men, which is about 4 times that of women globally (Chen W. et al., 2016; Bray et al., 2018). lncRNA Xist has recently been reported to regulate bladder cancer development through regulating several miRNAs or other target genes. lncRNA Xist exerts an oncogenic role through binding to miR-124, miR-139-5p, miR-200c, miR-133a, and miR-335 targets AR, Wnt1, TET1, and p53, which affect cell growth, invasion and migration, and metastasis (Hu et al., 2017; Xiong et al., 2017; Xu R. et al., 2018; Hu B. et al., 2019; Zhou et al., 2019c; Chen D. et al., 2020). This research uncovered that lncRNA Xist may be invoked as a potential therapeutic and prognostic biomarker for bladder cancer.
LncRNA Xist in Breast Cancer
Breast cancer accounts for almost one in four cancer cases among women, with respective incidence and mortality rates of 24.2 and 15.0%, and is the most commonly diagnosed cancer and leading cause of cancer death in women globally (Chen W. et al., 2016; Bray et al., 2018). Some previous studies have suggested that deregulation of lncRNA Xist plays a vital role in the pathogenesis of both inherited and sporadic breast cancer (Kawakami et al., 2004; Soudyab et al., 2016). The Breast Cancer 1 protein (BRCA1) is a tumor suppressor. Reduced expression of BRCA1 leads to increased risk of breast cancer development (Romagnolo et al., 2015). LncRNA Xist, which is dependent on the production of BRCA1 and may participate in regulating breast cancer development, is highly expressed in BRCA1-like breast cancer as a predictive biomarker (Sirchia et al., 2005, 2009; Vincent-Salomon et al., 2007; Schouten et al., 2016). It is thought that histone modifications (histone deacetylase inhibitor) and DNA methylation plays a critical role in breast cancer growth and metastasis (Librizzi et al., 2015; Shukla et al., 2019). Some research indicated that breast tumors frequently display major epigenetic instability of XI which is mediated by lncRNA Xist, and this phenomenon regulates breast cancer cells’ proliferation and differentiation (Salvador et al., 2013; Chaligne et al., 2015). In addition to the indirect regulation of competing endogenous RNA (ceRNAs), studies published to date have demonstrated that knockdown or overexpressed LncRNA Xist in breast cancer results in sponging five miRNAs, containing miR-155, miR-20a, miR-200c-3p, miR-125b-5p, and miR-362-5p, and positively regulates the downstream targets including CDX1, TP53, ANLN, NLRC5, and UBAP1, which affects breast cancer cells’ growth, proliferation, metastasis, migration, invasion, apoptosis, epithelial mesenchymal transition (EMT), and doxorubicin resistance (Zhao L. et al., 2018; Zheng R. et al., 2018; Li et al., 2020e; Liu et al., 2020a; Zhang et al., 2020a).
Triple-negative breast cancer (TNBC) is a subtype of breast cancer that accounts for approximately 10–20% of total breast cancer cases (Prat et al., 2015; Bianchini et al., 2016; Medina et al., 2020). The deficiency of estrogen, progesterone, and ERBB2 receptor expression leads to its highly invasive nature and relatively low response to current therapeutics approaches. Collectively, lncRNA Xist interacts with miR-454 to inhibit cell growth in TNBC (Li et al., 2020d). And lncRNA Xist sponges with miR-92b/Slug/ESA signaling pathway to suppresses TNBC growth (Li et al., 2020c). lncRNA Xist also positively regulates PHLPP1 expression via sequestering HDAC3 from the PHLPP1 promoter to influence cells’ viability (Huang et al., 2016). In cancer immunity and brain metastasis, lncRNA Xist involves cancer immunity in high expression programmed cell death protein 1 ligand TNBC cells via activating both OCT4 and NANOG though activating PI3K/AKT/mTOR signaling pathway (Salama et al., 2019). lncRNA Xist also promotes brain metastasis in breast cancer by activating the MSN-c-Met pathway and augmenting secretion of exosomal miR-503 (Xing et al., 2018), which may serve as an effective target for the treatment of brain metastasis. These findings demonstrate LncRNA Xist may contribute to a significant approach to the treatment of breast cancer.
LncRNA Xist in Colorectal Cancer
Colorectal cancer, with respective incidence and mortality rates of 10.2 and 9.2% in the world and which presents a rising trend in recent decades in China, ranks third in term of incidence but second in terms of mortality (Chen W. et al., 2016; Bray et al., 2018). As previously mentioned, lncRNA Xist exerts its function in colorectal cancer cells’ development by serving as a miRNA sponge. Zhang et al. (2019e) reported that lncRNA Xist, which modulates tumor size, plays a critical role in clinical prognosis and progression of colorectal cancer. Growing evidence from recent studies has shown that lncRNA Xist facilitates proliferation, metastasis, invasion, and EMT of colorectal cancer cells by functioning as an endogenous sponge of miR-200b-3p, miR-137, miR-132-3p, miR-486-5p, and miR-93-5p, thus affecting the expression of miRNAs target gene containing ZEB1, EZH2, MAPK1, NRP-2, and HIF-1A (Chen D.L. et al., 2017; Song et al., 2017; Liu et al., 2018a, 2019a; Yang L.G. et al., 2020). But beyond that, lncRNA Xist has been identified as the downstream target of methyltransferase-like14 (METTL14) by RNA-seq and Me-RIP, and its expression negatively correlating with METTL14 and YTHDF proteins 2 (YTHDF2) has been observed in colorectal cancer tissues (Yang X. et al., 2020). Yang X. et al. (2020) identified that METTL14-YTHDF2-lncRNA Xist axis mediated cells’ proliferation and metastasis in colorectal cancer.
In drug resistance of colorectal cancer cells, lncRNA Xist has been implicated in the resistance of colorectal cancer cells to chemoresistance via serving as a miRNA sponge. lncRNA Xist participates in the processes of drug resistance by modulating the axis of miR-124/serum and SGK1, miR-338-3p/PAX5, and miR-30a-5p/ROR1 (Zhu J. et al., 2018; Zhang et al., 2019d; Ma et al., 2020). Interestingly, Xiao et al. (2017) reported that overexpression of lncRNA Xist in colorectal cancer confers a potent poor therapeutic efficacy, and lncRNA Xist enjoys 5FU resistance via enhancing the expression of thymidylate synthase. In summary, this information indicates that lncRNA Xist may serve as an independent risk factor for colorectal cancer prognosis, and could be a potential therapeutic target and prognostic biomarker for colorectal cancer patients (Yu J. et al., 2020).
LncRNA Xist in Glioblastoma
Glioblastoma (GBM), with incidence rates of 3.2 per 100,000 and relative 5-year mortality rate of just 94.9%, is the most common and lethal primary intracranial tumor with few advances in treatment over the last several decades (Batash et al., 2017; McFaline-Figueroa and Wen, 2017; Kim et al., 2018). Accumulating evidence suggests that lncRNA Xist has a pivotal role in regulating glioma cells’ properties by interacting with miRNA. lncRNA Xist affects glioblastoma development by directly binding miR-152 and miR-429 (Yao et al., 2015; Cheng Z.H. et al., 2017). However, the downstream target gene of miR-152 and miR-429 remains unclear. In addition, lncRNA Xist mediates glioma progression, tumorigenesis, metastasis, proliferation, apoptosis, and glucose metabolism by positively regulating Bcl-2, FOXC1, ZO-2, Rac1, ASCT2, SLC1A5, SOX4, Smurf1, and IRS1 by functioning as a ceRNA of miR-204-5p, miR-137, miR-133a, miR-27a, and miR-126 (Wang Z. et al., 2017; Yu H. et al., 2017; Cheng Z.H. et al., 2020; Luo C.X. et al., 2020; Shen J. et al., 2020; Sun Y. et al., 2020; Yao et al., 2020; Zhao Q. et al., 2020). In drug resistance of glioblastoma cells, lncRNA Xist has been demonstrated in the resistance of human glioblastoma cells to Temozolomide (TMZ) via the miR-29c/DNA mismatch repair (MMR) pathway (Du P. et al., 2017). And Velázquez-Flores et al. (2020) have reported that XIST and XIST-210 may act as potential biomarkers for Diffuse intrinsic pontine gliomas diagnosis and prognostic biomarkers. In summary, these findings revealed that lncRNA Xist has an oncogenic role in the tumorigenesis of glioma and may serve as a novel and potential therapeutic target for patients with glioblastoma.
LncRNA Xist in Hepatocellular Carcinoma
Liver cancer, with respective incidence and mortality rates of 4.7 and 8.2%, was predicted to be the sixth most commonly diagnosed cancer and the fourth leading cause of cancer death worldwide in 2018 (Chen W. et al., 2016; Bray et al., 2018). The major risk factors of hepatocellular carcinoma are chronic infection with hepatitis B virus (HBV) or hepatitis C virus (HCV), aflatoxin-contaminated foodstuffs, heavy alcohol intake, obesity, smoking, and type 2 diabetes, and accounts for about 75–85% of primary live cancer (Chen W. et al., 2016; Bray et al., 2018). Recent studies have proposed that lncRNA Xist exerts tumorigenesis in hepatocellular carcinoma (Ma W.J. et al., 2017; Ma X. et al., 2017). LncRNA Xist, which functions as a ceRNA to regulate target HMGB1, SOX6, Smad7, PDK1/AKT, MAPK1, PDCD4, and PTEN expression by sponging miR-29b, miR-155-5p, miR-92b, miR-139-5p, miR-194-5p, miR-497-5p, and miR-181a, facilitates cells’ growth, autophagy, metastasis, and invasion via activating the miRNA/target signaling pathway (Zhuang et al., 2016; Chang et al., 2017; Mo et al., 2017; Kong et al., 2018; Lin et al., 2018; Xie et al., 2019; Zhang et al., 2019h). Analogously, Liu and Xu (2019) also demonstrated that silencing lncRNA Xist, whose expression level is significantly higher in hepatocellular carcinoma tissue compared with adjacent tissues, inhibits cell growth and tumor formation in hepatocellular carcinoma by directly interacting with miR-200b-3p, but the downstream target gene of miR-200b-3p remains unclear. All in all, these studies will contribute to providing a promising treatment for hepatocellular carcinoma.
LncRNA Xist in Nasopharyngeal Carcinoma
Nasopharyngeal carcinoma, with incidence rates of 0.7% and unknown mortality rates, is relatively uncommon compared with other cancers and is one of the most common malignant tumors in the head and neck (Chua et al., 2016; Wei K.R. et al., 2017). Accumulating studies suggests that the molecular function of lncRNA Xist has a pivotal function in nasopharyngeal carcinoma properties, such as cell proliferation, migration, and invasion. Knockdown of lncRNA Xist, which negatively regulates expression of miR-29c and miR-491-5p whose target gene remains unclear, suppressed cell proliferation, invasion, and growth and induces apoptosis in nasopharyngeal carcinoma (Han et al., 2017; Cheng Q. et al., 2018). Analogously, lncRNA Xist, which is highly expressed in nasopharyngeal carcinoma tissues and cell lines, facilitates nasopharyngeal carcinoma development via activating miR-34a-5p/E2F3, miR-148a-3p/ADAM17, and miR-381-3p/NEK5 axis (Song et al., 2016; Shi et al., 2020; Zhao C.H. et al., 2020). In drug resistance of nasopharyngeal carcinoma cells, lncRNA Xist, which may present a novel and potential therapeutic target in nasopharyngeal carcinoma, has been implicated in the resistance of human nasopharyngeal carcinoma cells to cisplatin (DDP) by facilitating programmed cell death 4 (PDCD4) and Fas ligand (Fas-L) expression (Wang et al., 2019b). On the whole, these reports will be play a novel role in the treatment of nasopharyngeal carcinoma.
LncRNA Xist in Lung Cancer
Lung cancer, with respective incidence and mortality rates of 11.6 and 18.4%, is the second most common cancer and remains the leading cause of cancer incidence and mortality worldwide (Chen W. et al., 2016; Bray et al., 2018). Emerging research demonstrates that lncRNA Xist are usually dysregulated in lung cancer and play a pivotal function in lung carcinoma initiation, progression, and therapy. lncRNA Xist, which has an oncogenic role in lung carcinoma, is closely correlated with tumor progression via regulating miR-140/iASPP axis and TCF-4 expression (Tang et al., 2017; Sun and Xu, 2019). Lung adenocarcinoma, which account for approximately 40% of total lung carcinoma, is also the most common histological subtype of NSCLC (Rong et al., 2020). lncRNA Xist expedites cancer progression and the resistance of cisplatin in lung adenocarcinoma via mediating the miR-363-3p/MDM2 and let-7i/BAG-1 signaling pathway (Sun J. et al., 2017; Rong et al., 2020). These results indicated that lncRNA Xist is likely to be a new marker and potential therapeutic target for patients with lung adenocarcinoma.
Non-small cell lung carcinoma (NSCLC), which accounts for 85% of lung cancer cases, is the most common subtype of lung cancer (Zhou et al., 2018). Accumulating evidence has revealed that lncRNA Xist is a pivotal regulator of cell proliferation, EMT, migration, invasion, and drug resistance in NSCLC. lncRNA Xist acts as an oncogene in NSCLC by modulating HIF1A-AS1 and KLF2 expression (Tantai et al., 2015; Fang et al., 2016). lncRNA Xist also positively mediates Bcl-2, LARP1, PXN and Notch-1, CBLL1, and RING1 expression by functioning as a ceRNA of miR-449a, miR-374a, miR-137, miR-212-3p, and miR-744, which are involved in cell proliferation, migration, invasion, EMT, and death in NSCLC (Xu Z.Z. et al., 2017; Zhang Y.L. et al., 2017; Jiang H.J. et al., 2018; Wang et al., 2018c, 2019c; Qiu et al., 2019). In addition, lncRNA Xist (Wang H.Y. et al., 2017), which has a higher expression in NSCLC cell lines and tissues, increases cell proliferation and invasion by negatively regulating miR-186-5p expression; however, the downstream target gene of miR-186-5p remains unclear.
It has been reported (Li C. et al., 2018) that TGF-β (Transforming growth factor β)-induced EMT serves a vital role in NSCLC metastasis and invasion. lncRNA Xist promotes TGF-β-induced EMT by positively regulating ZEB2 via interacting with miR-367 and miR-141 (Li C. et al., 2018). Analogously, lncRNA Xist inhibits NSCLC progression by sponging miR-16, miR-335, and miR-142-5p, and regulating target CDK8, SOD2/ROS, and PAX6 expression (Liu et al., 2019b; Zhou et al., 2019e; Jiang et al., 2020). Drug resistance is one of the most common reasons for therapeutic failure in patients with NSCLC and a persistent issue that requires continued investigation. Emerging evidence indicated that lncRNA Xist is associated with cisplatin resistance in NSCLC by TGF-β effector SMAD2 signaling pathway, miRNA-144-3p/MDR1 and MRP1, and miR-17/ATG7 axis (Sun W. et al., 2017; Tian et al., 2019; Xu et al., 2020). All in all, this evidence suggests that lncRNA Xist may offer a hopeful diagnostic and therapeutic choice for the treatment of NSCLC.
LncRNA Xist in Osteosarcoma
Bone cancer, with respective incidence and mortality rates of 0.20 and 0.28%, occurs frequently in children, adolescents, and young adults aged 15 to 29 years (Siegel et al., 2019, 2020). Osteosarcoma, which accounts for 20 to 40% of all bone tumors, are the most frequent morphological subtypes of bone cancer, representing a worldwide and common primary malignant bone tumor in children and adolescents (Balmant et al., 2019; Muller and Silvan, 2019). Growing evidence from recent studies has shown that lncRNA Xist is aberrantly regulated in osteosarcoma. LncRNA Xist, which participated in osteosarcoma development processes, including cell proliferation, migration, invasion, EMT, and apoptosis, is involved in gene regulation through a variety of mechanisms, primarily by functioning as a miRNA sponge and via interacting with its targets (Li et al., 2017; Wang et al., 2019f; Han and Shen, 2020), such as miR-153/SNAI1 pathway (Wen et al., 2020), EZH2, PUMA, and NF-kB (Xu T. et al., 2017; Gao et al., 2019).
In addition to indirect modulation of ceRNAs, studies published to date have indicated that high lncRNA Xist expression in osteosarcoma results in sponging six miRNAs, namely miR-21-5p, miR-193a-3p, miR-195-5p, miR-320b, miR-137, and miR-375-3p, which affects osteosarcoma progression (Wu D.P. et al., 2017; Zhang and Xia, 2017; Lv et al., 2018; Yang C. et al., 2018; Li H. et al., 2019; Sun X. et al., 2019). lncRNA, which regulates miR-21-5p/PDCD4 axis, miR-193a-3p/RSF1 axis, miR-195-5p/YAP axis, miR-137, miR-320b/RAP2B axis, and miR-375-3p/KT/mTOR axis, contributes to osteosarcoma cell growth, metastasis, and invasion by activating MAPK signaling pathway, NF-kB signaling pathway, and PI3K-AKT-mTOR signaling pathway. Taking all into account, these studies indicated that lncRNA Xist may act as a candidate prognostic biomarker and a promising therapeutic target for osteosarcoma (Wu D.P. et al., 2017; Zhang and Xia, 2017; Lv et al., 2018; Yang C. et al., 2018; Li H. et al., 2019; Sun X. et al., 2019).
LncRNA Xist in Pancreatic Cancer
Pancreatic cancer, with respective incidence and mortality rates of 2.5% (China, 2.1%, 2015) and 4.5% (China, 2.8%, 2015), was the seventh leading cause of cancer death worldwide in both males and females in 2018 (Chen W. et al., 2016; Bray et al., 2018). Accumulating evidence indicated that lncRNA Xist interacts with additional miRNAs, such as miR-133a, miR-140 and miR-124, miR-34a-5p, miR-34a, miR-141-3p, and miR-429 in pancreatic cancer, and is involved in the development and progression of pancreatic cancer (Liang et al., 2017; Wei W. et al., 2017; Sun Z. et al., 2018; Shen et al., 2019; Sun and Zhang, 2019; Zou et al., 2020). As aforementioned, lncRNA Xist promotes pancreatic cancer cells’ proliferation by binding miR-133a, thus affecting the miR-133a downstream target gene EGFR (epidermal growth factor receptor) which is positively correlated with lncRNA Xist (Wei W. et al., 2017). lncRNA Xist also facilitates miR-140/miR-124/iASPP/CDK1 axis, miR-34a/YAP axis, miR-141-3p/TGF-β2 axis, miR-429/ZEB1 axis and miR-34a-5p, which contributes to carcinoma cell growth, EMT, migration, and invasion (Liang et al., 2017; Sun Z. et al., 2018; Shen et al., 2019; Sun and Zhang, 2019; Zou et al., 2020). However, the downstream target gene of miR-34a-5p remains unknown. Taken together, the above research results suggested that lncRNA Xist could be regarded as a candidate prognostic biomarker and a potential therapeutic target in human pancreatic carcinoma.
LncRNA Xist in Retinoblastoma
Retinoblastoma, which has a significant effect on mortality in emerging countries but is more curable in industrialized countries, is an aggressive eye cancer that affects infants and children (Cassoux et al., 2017). Recently, abundant studies demonstrated that dysregulation lncRNA was involved in tumorigenesis and cancer progression of retinoblastoma (Yang and Wei, 2019). Compared to healthy controls, lncRNA Xist was significantly upregulated in plasma of retinoblastoma patients which was inversely associated with lncRNA NKILA (Lyu et al., 2019). LncRNA Xist overexpression promotes retinoblastoma cells proliferation, migration, and invasion rates via negatively regulating lncRNA NKILA, but the causality has not been fully validated. In addition, lncRNA Xist, which indirectly interacts with miR-21-5p, miR-124, miR-101, miR-140-5p, and miR-200a-3p, and positively regulates VEGF, STAT3, ZEB1 and ZEB2, SOX4, and NRP1 expression, facilitates apoptosis, migration, EMT, proliferation, and invasion by activating signaling pathways, such as PI3K-Akt signaling pathway and MAPK-ERK signaling pathway (Hu et al., 2018; Cheng Y. et al., 2019; Dong et al., 2020; Wang et al., 2020c; Zhao H. et al., 2020). All in all, these studies suggested that lncRNA Xist serves a potential and promising clinical application for diagnosis, prognosis, and treatment.
LncRNA Xist in Cervical Cancer
Cervical cancer, with respective incidence and mortality rates of 3.2 and 3.3%, ranked as the fourth most frequently diagnosed cancer and the fourth leading cause of cancer death in women in 2018 worldwide (Chen W. et al., 2016; Bray et al., 2018). Recently, numerous reports found that dysregulation of lncRNA Xist was involved in regulating cervical cancer progression via binding to miRNAs. Zhu H. et al. (2018) demonstrated lncRNA Xist, which is extremely highly expressed in cervical cancer tissues and cell lines, accelerates cervical cancer progression via upregulating Fus through functioning as a ceRNA of miR-200a. In additional, lncRNA Xist upregulation, which positively facilitates ORC1 expression and acts as a ceRNA of miR-140-5p, contributes to the cervical cancer progression by activating miR-140-5p/ORC1 axis (Chen X. et al., 2019c). Similarly, Liu et al. (2020d) found that lncRNA Xist, which was highly expressed in cervical cancer cells and tissue, promoted cervical cancer cells’ proliferation, migration, and invasion and hindered apoptosis by inhibiting miR-889-3p and positively mediating SIXI expression. Taken together these studies demonstrated that lncRNA Xist may play a role in epigenetic diagnostics and therapeutics in cervical cancer.
LncRNA Xist in Gastric Cancer
Stomach cancer (cardia and non-cardiac gastric cancer combined), with respective incidence and mortality rates of 5.7 and 8.2%, was the fifth most frequently diagnosed cancer and the third leading cause of cancer death in 2018 worldwide, and remains an important cancer (Chen W. et al., 2016; Bray et al., 2018). Recently, some reports founded that lncRNA Xist exerts its function in gastric cancer progression by acting as a miRNA sponge, It acts on miRNA, such as miR-101, miR-497, miR-185, and miR-337. lncRNA Xist, which acts as a molecular sponge of miR-101, miR-497, miR-185, and miR-337 to mediate EZH2, MACC1, TGF-β1, and JAK2 expression, is involved in gastric cancer progression through mediating miR-101/EZH2 axis, miR-497/MACC1 axis, miR-185/TGF-β1 axis, and miR-337/JAK2 axis (Chen D.L. et al., 2016; Ma L. et al., 2017; Zhang Q. et al., 2018; Zheng W. et al., 2020). In drug resistance of gastric carcinoma cells, Li Y.D. et al. (2019) demonstrated that lncRNA Xist contributes to drug resistance of gastric cancer cells though positively facilitating the related gene MDR1 (multidrug resistance gene 1) and MRP1 (multi-drug resistance protein 1) of multidrug resistance, and is helpful for the molecule-targeted treatment of gastric cancer. Taken together, these findings suggest that lncRNA Xist may be a candidate prognostic biomarker and a new therapy target in gastric cancer patients.
LncRNA Xist in Melanoma
Melanoma, with respective incidence and mortality rates of 1.6 and 0.6%, was the most fatal form of skin cancer in 2018 worldwide and the rates are increasing faster than any other currently preventable cancers (Chen W. et al., 2016; Bray et al., 2018; Schadendorf et al., 2018). Recently, some findings suggested that a major role of lncRNA Xist is facilitating melanoma progression via acting as a miRNAs sponge to regulate its downstream target genes, such as lncRNA Xist (Zhang et al., 2019f), which promoted malignant melanoma growth and metastasis by functioning as a ceRNA though miR-217. However, the downstream target gene of miR-217 remains unclear. Analogously, lncRNA Xist (Pan et al., 2019; Tian K. et al., 2020), which functions as a ceRNA to positively regulate ROCK1 and PI3KRI and AKT expression by sponging miR-139-5p and miR-21, respectively, facilitates proliferation, invasion, and oxaliplatin resistance of melanoma cells. In summary, this evidence shows that lncRNA Xist could provide a novel insight into the pathogenesis and underlying therapeutic targets for melanoma.
LncRNA Xist in Esophageal Cancer
Esophageal cancer, with respective incidence and mortality rates of 3.2 and 5.3%, ranks seventh in terms of incidence and sixth in mortality overall, the latter signifying that esophageal cancer was responsible for an estimated 1 in every 20 cancer deaths in 2018 worldwide (Chen W. et al., 2016; Bray et al., 2018; Schadendorf et al., 2018). Recently, numerous reports demonstrated that dysregulation of lncRNA Xist was involved in regulating esophageal cancer development via binding to miRNAs. lncRNA Xist involves esophageal squamous cell carcinoma development via regulation of miR-101/EZH2 axis (Wu X. et al., 2017), and facilitates esophageal squamous cell carcinoma proliferation, apoptosis, migration, and invasion via regulation miR-494/CDK6 axis and activation of JAK2/STAT3 signal pathway (Chen et al., 2019f). In addition, lncRNA Xist predicts the presence of lymph node metastases in human esophageal squamous cells (Li and He, 2019; Wang et al., 2020b). Taken together, these results demonstrated that lncRNA Xist may provide a novel candidate prognostic biomarker and a new insight for esophageal carcinoma therapy.
LncRNA Xist in Laryngeal Squamous Cell Carcinoma
Laryngeal cancer, with respective incidence and mortality rates of 1.0 and 1.0%, was one of the most common tumors of the respiratory tract in 2018 worldwide (Chen W. et al., 2016; Steuer et al., 2017; Bray et al., 2018). Squamous cell carcinoma, accounting for approximately 90% of malignant neoplasms of the larynx, is the most common malignancy of the larynx (Thompson, 2017; Bradford et al., 2020). Recently, some evidence suggested that a major role of lncRNA Xist, which is notably up-regulated in laryngeal squamous cell carcinoma tissues and cells, promotes laryngeal squamous cell carcinoma progression via interacting with miRNAs to regulate its downstream target gene. lncRNA Xist increases the aggressiveness of laryngeal squamous cell carcinoma by functioning as a ceRNA sponge of miR-124 to regulate EZH2 expression (Xiao D. et al., 2019), and promotes progression of laryngeal squamous cell carcinoma via activating the miR-144/IRS1 axis (Cui et al., 2020), and promotes the malignance of laryngeal squamous cell carcinoma cells through functioning as a ceRNA of miR-125b-5p to positively modulate TRIB2 expression (Liu et al., 2020b). All together, these studies demonstrated that lncRNA Xist may serve as a new potential prognostic biomarker and putative target in the therapy of laryngeal squamous cell carcinoma.
LncRNA Xist in Ovarian Cancer
Ovarian cancer, which is the leading cause of death for women of reproductive age around the world and has a 5-year survival rate below 45%, is in eighth place among the most common cancers in women and the fifth leading cause of death among women worldwide, including 4% of all cancers (Webb and Jordan, 2017; Moga et al., 2018; Stewart et al., 2019). In order to prove the lncRNA Xist participated in ovarian cancer development, Wang et al. (2018a) revelated that lncRNA Xist is involved in ovarian cancer development by negatively regulating miR-214-3p expression. This confirmed that lncRNA Xist is closely associated with the tumor grade and distant metastasis in the ovarian cancer patients (Zuo et al., 2019). This result suggested that lncRNA Xist plays a role in tumor development. In addition, lncRNA Xist (Wang and Li, 2020), which functions as a ceRNA to positively mediate the expression of PDCD4 (programmed cell death protein 4) through binding to miR-150-5p and is significantly decreased in ovarian cancer tissues and cell lines compared with the normal tissue and cells, inhibits ovarian cancer cell growth and metastasis via regulating miR-150-5p/PDCD4 signaling pathway. All in all, these studies evaluated that lncRNA Xist provides insight into the potential target for the treatment of ovarian cancer, and a new evaluation of the diagnosis and prognosis of ovarian cancer.
LncRNA Xist in Others Cancer
Growing evidence from recent studies has shown that lncRNA Xist facilitates tumor development, including pituitary neuroendocrine tumor, neuroblastoma, thyroid cancer, colon cancer, renal cell carcinoma, and prostate cancer (Chaligne and Heard, 2014; Yang Z. et al., 2018; Liu et al., 2019c). In the pituitary neuroendocrine tumor cells (Zhou et al., 2019b), lncRNA Xist, which functions as a ceRNA to sequester miR-424-5p to elevate the expression of the its target bFGF, and exhibits high expression in invasive pituitary neuroendocrine tumor tissues as compared to non-invasive tumor tissues, promotes cancer progression in invasive pituitary neuroendocrine tumor via activating the miR-424-5p/bFGF signaling pathway. In neuroblastoma (Zhang et al., 2019a), lncRNA Xist, which interacts with EZH2 to downregulate DKK1 by inducing H3 histone methylation, promotes neuroblastoma cell growth, proliferation, migration, and invasion via modulating H3 histone methylation of DKK1 in neuroblastoma. In addition, lncRNA Xist (Yang H. et al., 2020) repressed tumor growth and boosted radiosensitivity of neuroblastoma via modulating the miR-375/L1CAM axis. In thyroid cancer, lncRNA Xist (Liu et al., 2018c), which positively regulates MET by sponging miR-34a, modulates the cell proliferation and tumor growth through activating the PI3K/AKT signaling pathway. Analogously, Xu Y. et al. (2018) demonstrated that lncRNA Xist, whose high expression is positively associated with TNM stage and lymph node metastasis, promotes cell proliferation and invasion by interacting with miR-141 in papillary thyroid carcinoma. However, the downstream target gene of miR-141 remains unknown. In colon cancer cells, lncRNA Xist (Sun N.N. et al., 2018), which functions as a ceRNA by binding to miR-34a and positively modulates WNT1 expression, has a crucial function in colon cancer progression via the miR-34a/WNT1 axis to activate the Wnt/β-catenin signaling pathway.
In addition to indirect regulation of ceRNAs, studies published to date have manifested that low lncRNA Xist expression in renal cell carcinoma results in sponging two miRNAs, miR-106b-5p and miR-302c, which regulates tumor development (Zhang J. et al., 2017; Sun K. et al., 2019). lncRNA Xist, which positively facilitates P21 and SDC1 expression through sponging miR-106b-5p and miR-302c, facilitates cell proliferation and apoptosis via miR-106b-5p/P21 signaling pathway and miR-302c/SDC1 axis. In prostate cancer cells, lncRNA Xist, which weakly expresses in normal prostate tissues but not in leukocytes, contributes prostate cancer development (cell proliferation and metastasis) by activating miR-23a/RKIP signaling pathway (Laner et al., 2005; Du Y. et al., 2017). In addition, SQ. Hai (2020) have indicated that LncRNA XIST/miR-124-3p/iASPP Pathway Promotes Growth of Human Chordoma Cells. Analogously, Lobo et al. (2019) manifested that demethylated and methylated XIST promoter may be involved in testicular germ cell tumor development. Altogether, this evidence pronounced that lncRNA Xist may shed new light on epigenetic diagnostics and therapeutics for cancer patients.
The Role of LncRNA Xist in Non-Cancer Diseases
Diseases are abnormal conditions that have a specific set of signs and symptoms. Diseases can have an external cause, such as an infection, or an internal cause, such as autoimmune disease (such as Alzheimer’s disease). Accumulating evidence has suggested that lncRNA Xist participates in non-cancer related diseases’ development and progression (Table 2) as a ceRNA regulatory network of miRNA-mRNA. It also provides a window into the understanding of aberrant expression of lncRNA Xist associated with non-cancer related diseases. lncRNA Xist is a novel potential biomarker and could potentially be involved in the diagnosis and therapy of different types of diseases.
TABLE 2.
LncRNA Xist and miRNA in non-cancer related disease.
| Disease type | miRNA | Target | Mechanism of action and function | Signaling pathway | References |
| Cardiac disease | miR-330-3p, miR-101 | S100B, TLR2 | Aberrant expression lncRNA Xist is involved in non-cancer related diseases and cells development, such as cell apoptosis, cell cycle, cell proliferation, cell differentiation. | TGF-beta signaling pathway, PI3K-Akt signaling pathway, Toll-like receptor signaling pathway, cAMP signaling pathway, Notch signaling pathway, Prolactin signaling pathway, JAK-STAT signaling pathway, Toll-like receptor signaling pathway, NF−κB signaling pathway, NOD-like receptor signaling pathway, C-type lectin receptor signaling pathway, Hedgehog signaling pathway, Thyroid hormone signaling pathway, HIF-1 signaling pathway, Wnt/β-catenin signaling pathway, BMP/TGF-β signaling pathway, MAPK and MMPs signaling pathway, Human papillomavirus infection, AGE-RAGE signaling pathway in diabetic complications, Relaxin signaling pathway, T cell receptor signaling pathway and B cell receptor signaling pathway, etc. | Chen Y. et al., 2018; Xiao L. et al., 2019 |
| Myocardial infarction | miR-130a-3p, miR-101a-3p | PDE4D, FOS | Zhou et al., 2019d; Lin B. et al., 2020 | ||
| Acute myocardial infarction | miR-449, miR-122-5p, miR-125b, miR-133a, miR-150-5p | Notch1, FOXP2, hexokianse 2, SOCS2, Bax | Li Z.Q. et al., 2019; Zhang et al., 2019c; Fan et al., 2020; Peng et al., 2020; Zhou et al., 2020 | ||
| Neuropathic pain | miR-133b-3p, miR-154-5p, miR-137, miR-544, miR-150 | Trisomy 21, Pitx3, TLR5, TNFAIP1, STAT3, ZEB1 | Jin et al., 2018; Yan et al., 2018; Zhao Y. et al., 2018; Wei et al., 2019 | ||
| Neurodegeneration | miR-133b-3p | Pitx3 | He et al., 2020 | ||
| Alzheimer’s disease | miR-132, miR-124 | BACE1 | Wang et al., 2018b; Du Y. et al., 2020 | ||
| Osteoarthritis | miR-211, miR-214-3p, miR-17-5p, miR-1277-5p, miR-376c-5p, miR-142-5p, miR-149-5p, miR-675-5p | CXCR4, MAPK, AHNAK, BMP2, TIMP-3, MMP-13, ADAMTS5, OPN, SGTB, DNMT3A, GNG5 | Li L. et al., 2018; Liao et al., 2019; Wang et al., 2019e; Feng et al., 2020; Ghaderian et al., 2020; Li et al., 2020b; Liu et al., 2020e; Shen X.F. et al., 2020 | ||
| Bone marrow | miR-9-5p | ALPL, ALP, Bglap, Runx2 | Zheng C. et al., 2020 | ||
| Inflammation | miR-27a-3p, miR-34a, miR-30c-5p, miR-146a | NF−κB, NLRP3, Smurf1, YY1, PTEN, Nav1.7 | Shenoda et al., 2018; Sun W.B. et al., 2018; Hu W.N. et al., 2019; Zhao Q. et al., 2020 | ||
| Spinal cord injury | miR-27a, miR-494, miR-32-5p | Smurf1, PTEN, Notch-1 | Gu et al., 2017; Cheng X. et al., 2020; Zhao Q. et al., 2020 | ||
| Acute kidney injury | miR-15-5p, miR-212-3p, miR-122-5p, miR-142-5p | CUL3, ASF1A, BRWD1M, PFKFB2, PDCD4 | Xu et al., 2019; Cheng and Wang, 2020; Tang et al., 2020 | ||
| Nephropathy | miR-217, miR-93-5p, miR-485 | TLR4, CDKN1A, PSMB8 | Jin et al., 2019; Yang et al., 2019; Wang, 2020 | ||
| Placental angiogenesis | miR-429, miR-485-3p | VEGF-A, SOX7, ERK1/2, Akt | Chen G. et al., 2018; Hu C. et al., 2019 | ||
| Acute pneumonia | miR-370-3p | TLR4, JAK, STAT, NF−κB | Zhang et al., 2019g | ||
| Pulmonary fibrosis | miR-139 | β-catenin | Wang Y.C. et al., 2017 | ||
| Primary graft dysfunction | miR-21 | IL-12A | Li et al., 2020a | ||
| Rett syndrome | MeCP2, BMP/TGF-β | Sripathy et al., 2017; Adrianse et al., 2018 | |||
| Acute respiratory distress syndrome | miR-204 | IRF2 | Wang et al., 2019d | ||
| SCNT embryo development | REX1, YY1, MSL1/MSL2 | ||||
| Human trophoblast cells | miR-144 | Titin, MAPK, MMPs | Yu N.H. et al., 2017 | ||
| Endothelial cells injury | miR-320 | NOD2 | Xu X.H. et al., 2018 | ||
| Skin fibroblasts | miR-29a, miR-29b-3p | LIN28A, COL1A1 | Guo et al., 2018; Cao and Feng, 2019 | ||
| Osteoblasts | miR-203-3p, let-7c-5p | ZFPM2, STAT3 | Niu et al., 2020; Wang et al., 2020d | ||
| Keratoconus | miR-181a | COL4A1 | Tian R. et al., 2020 | ||
| Hair follicle regeneration | miR-424 | Shh | Lin B.J. et al., 2020 | ||
| Acute liver injury | BRD4 | Shen and Li, 2020 | |||
| Stanford Type A Aortic Dissection | miR-17 | PTEN | Zhang et al., 2020b |
LncRNA Xist in Cardiac Disease
Cardiac diseases, including coronary artery disease (CAD), myocardial infarction (MI), cardiac hypertrophy, and heart failure (HF), are among the leading causes of morbidity and mortality worldwide (Greco et al., 2018; Colpaert and Calore, 2019). Emerging evidence has revealed that lncRNA Xist acted as powerful and dynamic modifier of cardiac physiological and pathological processes. Cardiac hypertrophy, recognized as a risk predictor of sudden cardiac death, is an adaptive reaction in response to altered stress or injury to maintain cardiac function (Li Y. et al., 2018; Wehbe et al., 2019; Luo X. et al., 2020). lncRNA Xist (Sohrabifar, 2020) participates in the pathogenesis of complex diseases and also serves as a diagnostic marker. lncRNA Xist also positively regulates S100B expression through functioning as a ceRNA to bind miR-330-3p (Chen Y. et al., 2018) and functions as a ceRNA of miR-101 to enhance TLR2 expression (Xiao L. et al., 2019) and modulates the progression of cardiomyocyte hypertrophy by miR-330-3p/S100B pathway and miR-101/TLR2 axis.
Myocardial infarction (MI), colloquially known as “heart attack,” is caused by decreased or complete cessation of blood flow to a portion of the myocardium and by the rupture of atherosclerotic plaques, which results in damage to cardiomyocytes due to lack of oxygen (Colpaert and Calore, 2019; Ojha and Dhamoon, 2020). lncRNA Xist, which positively mediates PDE4D expression via interacting miR-130a-3p (Zhou et al., 2019d) and targets miR-101a-3p through regulating FOS (Lin B. et al., 2020), promotes myocardial infarction development and cell apoptosis, and inhibits cell proliferation though the miR-130a-3p/PDE4D aixs and miR-101a-3p/FOS aixs.
Acute MI (AMI) is characterized by ischemic injury and cardiomyocyte apoptosis, while myocardial injury, which is also an entity in itself, is a prerequisite for the diagnosis of MI in the setting of acute myocardial ischemia (Sandoval and Thygesen, 2017; Sandoval et al., 2017; Colpaert and Calore, 2019). lncRNA Xist, which interacts directly with miRNA (miR-150-5p, miR-122-5p, miR-125b, miR-133a, and miR-449) to positively regulate expression levels of mRNA (Bax, FOXP2, hexokianse 2, SOCS2, and Notch1), protects hypoxia-induced cardiomyocyte injury and represses the myocardial cell apoptosis though miR-150-5p/Bax pathway, miR-122-5p/FOXP2 axis, miR-125b/hexokianse 2 axis, miR-133a/SOCS2 pathway, and miR-449/Notch1 signaling pathway (Li Z.Q. et al., 2019; Zhang et al., 2019c; Fan et al., 2020; Peng et al., 2020; Zhou et al., 2020). These results indicated that lncRNA Xist represents a very promising potential pharmacotherapeutic target and biomarker for cardiac disease.
LncRNA Xist in Neuropathic Pain
Neuropathic pain, including central pain, peripheral pain, and cancer pain, is pain that arises as lesions or diseases of the somatosensory system, either at the peripheral or at the central level, and are treated by first-line (include antidepressants and anticonvulsants acting at calcium channels), second-, and third-line (include topical lidocaine and opioids) pharmacologicals (Xu et al., 2016; Fornasari, 2017; Eberlin, 2019). Growing studies have revealed that lncRNA Xist, which has been characterized as a key modulator of neuronal functions, plays a pivotal role in the development of neuropathic pain. In Down’s syndrome, lncRNA Xist (Czerminski and Lawrence, 2020), which fully corrects trisomy 21 dosage in neural cells, promotes differentiation of trisomic NSCs (neural stem cells) to neurons by silencing Trisomy 21 and activating Notch signaling pathway. In Parkinson’s disease (PD) animals, it has been shown that lncRNA Xist/miR-133b-3p/Pitx3 axis protect dopaminergic neurons through activation of CB2R with AM1241, which alleviates PD (He et al., 2020). In addition, lncRNA Xist participated in neuropathic pain though interacting with miRNAs in CCI (chronic constriction injury) rat models, including miR-154-5p, miR-137, miR-544, and miR-150. lncRNA Xist, which functions as a ceRNA to positively modulate mRNA expression (TLR5, TNFAIP1, STAT3, and ZEB1) by sponging miRNA (miR-154-5p, miR-137, miR-544, and miR-150), contributes to neuropathic pain development by facilitating miR-154-5p/TLR5 axis, miR-137/TNFAIP1 axis, miR-544/STAT3 axis, and miR-150/ZEB1 axis in CCI rat models (Jin et al., 2018; Yan et al., 2018; Zhao Y. et al., 2018; Wei et al., 2019).
Alzheimer’s disease (AD), which is a growing global health concern with huge implications for individuals and society, is a chronic progressive and irreversible neurodegenerative disorder (Scheltens et al., 2016; Lane et al., 2018; Chanda and Mukhopadhyay, 2020). Silencing lncRNA Xist (Wang et al., 2018b) attenuated Aβ(amyloid-beta peptide)25-35-induced toxicity, oxidative stress, and apoptosis in primary cultured rat hippocampal neurons by negatively mediating miR-132 expression. But the downstream target gene of miR-132 remains unclear. Similarly, Du Y. et al. (2020) showed that lncRNA Xist, which was significantly upregulated in hydrogen peroxide (H2O2)-induced AD mice models and in H2O2-treated N2a cells, is involved in Alzheimer’s disease development though positively regulating BACE1 expression by interacting with miR-124. These studies suggested that lncRNA Xist might provide novel therapeutic avenues for neuropathic diseases.
LncRNA Xist in Osteoarthritis
Osteoarthritis (OA), which is the most common joint disorder that affects one or several diarthrodial joints including small joints (such as those in the hand) and large joints (such as the knee and hip joints), is the most frequently diagnosed musculoskeletal disease and leads to functional decline and loss in quality of life (Kraus, 2014; Pereira et al., 2015). Accumulated evidence manifested that lncRNA Xist is associated with development and progression of OA. lncRNA Xist, which acts as a ceRNA of miR-211 to positively mediate miR-211-interacted CXCR4 expression, promotes the proliferation and apoptosis of OA through the miR-211/CXCR4 axis activating MAPK signaling pathway (Li L. et al., 2018). And lncRNA Xist (Liao et al., 2019), which positively regulates AHNAK expression to activate BMP2 Signaling Pathway by target with miR-17-5p, may influence Cervical Ossification of the PLL through facilitating of miR-17-5P/AHNAK/BMP2 axis. In periodontal ligament stem cells (PDLSCs), lncRNA Xist, which was elevated in osteogenic inducted PDLSCs, promoted Osteogenic Differentiation by negatively regulating the expression of miR-214-3p, but the downstream target gene of miR-214-3p remains unknown (Feng et al., 2020).
In osteoporosis (OP), lncRNA Xist (Chen et al., 2019a,d), which was highly expressed in the serum and monocytes of patients with OP, regulates osteoporosis through recruiting DNA methyltransferase and inhibiting bone marrow mesenchymal stem cell differentiation. In addition, a major role of lncRNA Xist is facilitating gene expression and affecting osteoarthritis development and progression via sponging to miRNAs. The lncRNA Xist/miR-9-5p/ALPL (Zheng C. et al., 2020) and lncRNA Xist/miR-1277-5p/MMP-13 and ADAMTS5 (Wang et al., 2019e) signaling pathway has been identified as a ceRNA regulatory network involved in osteoarthritis development. And other ceRNA regulatory networks have also been shown to contribute to the progression of Osteoarthritis, such as lncRNA Xist/miR-376c-5p/OPN signaling pathway (Li et al., 2020b), lncRNA Xist/miR-142-5p/SGTB signaling pathway (Ghaderian et al., 2020), lncRNA Xist/miR-149-5p/DNMT3A signaling pathway (Liu et al., 2020e), and lncRNA Xist/miR-675-3p/GNG5 signaling pathway (Shen X.F. et al., 2020). Increasing studies have shown that lncRNA Xist might act as a novel therapeutic target for OA patients.
LncRNA Xist in Inflammation
Inflammation, which is activated by inflammasomes that are innate immune system receptors and sensors that regulate the activation of caspase-1, is a protective immune response mounted by the evolutionarily conserved innate immune system in response to harmful stimuli, such as pathogens, dead cells, or irritants, and is tightly regulated by the host (Guo et al., 2015). Recent findings demonstrated the pivotal role of lncRNA Xist in the progression of the inflammatory response. NF-κB (nuclear factor-κB) signaling pathway, which plays a vital role in inflammation and innate immunity, were involved in cell proliferation and apoptosis and regulated the production of inflammatory cytokines including tumor necrosis factor (TNF)-α, interleukin (IL)-1β, IL−6, and IL−8 (Ma et al., 2019; Shenoda et al., 2020). lncRNA Xist facilitates acute inflammatory responses and bovine mammary epithelial cell inflammatory responses via NF-κB/NLRP3 inflammasome signaling pathway (Levey and James, 2017; Yang et al., 2019).
In addition to the indirect regulation of ceRNAs, studies published to date have demonstrated that high lncRNA Xist expression in inflammatory cells results in sponging four miRNAs, namely miR-27a-3p, miR-30c, miR-34c, and 146a, which responded to the inflammatory development process (Shenoda et al., 2018; Sun W.B. et al., 2018; Hu W.N. et al., 2019; Zhao Q. et al., 2020). The regulation of lncRNA Xist in most inflammation processes, including acute inflammation response in female cells, apoptosis, and inflammatory injury of microglia cells after spinal cord injury, cell apoptosis of HUVEC (Human umbilical vein endothelial cells) and ox-LDL (oxidized low-density lipoprotein)-induced the inflammatory response and inflammatory pain in rat. The pathways involved included lncRNA Xist/miR-34a/YY1 signaling pathway (Shenoda et al., 2018), lncRNA Xist/miR-27a/Smurf1 signaling pathway (Zhao Q. et al., 2020), lncRNA Xist/miR-30c-5p/PTEN signaling pathway (Hu W.N. et al., 2019), and miR-146a/Nav1.7 signaling pathway (Sun W.B. et al., 2018). These results suggest that lncRNA Xist could be involved in a promising strategy against inflammation and be a potential target for inflammatory patients.
LncRNA Xist in Kidney and Cardiovascular Disease
As the kidney and heart are intricately linked, abnormal function in one can lead to pathological function in the other (Lorenzen and Thum, 2016). Acute kidney injury (formerly known as acute renal failure), which is typically diagnosed by the accumulation of end products of nitrogen metabolism (urea and creatinine) or decreased urine output, or both, is a syndrome characterized by the rapid loss of the function of glomerular filtration rate (Levey and James, 2017; Ronco et al., 2019). Recent studies indicated lncRNA Xist exerts its function by serving as a miRNA sponge in the development of kidney injury. In diabetic nephropathy, lncRNA Xist, which is highly expressed in the kidney tissue of diabetic nephropathy mice and high glucose-exposed HK-2 cells, is involved in diabetic nephropathy development by positively facilitating CDKN1A (cyclin-dependent kinase inhibitor 1A) expression via functioning as a ceRNA of miR-93-5p (Yang et al., 2019). In LPS-induced SCI (Spinal cord injury) microglia cells and lncRNA Xist, which interacts with miR-27a to mediate the downstream target gene of Smurf1 expression, alleviated the apoptosis and inflammatory injury of microglia cells after SCI through activating miR-27a/Smurf1 axis (Zhao Q. et al., 2020). These signaling pathways, which include lncRNA Xist/miR-494/PTEN/PI3K/AKT signaling pathway (Gu et al., 2017), lncRNA Xist/miR-142-5p/PDCD4 signaling pathway (Tang et al., 2020), lncRNA Xist/miR-217/TLR4 signaling pathway (Jin et al., 2019), lncRNA Xist/miR-32-5p/Notch-1 signaling pathway (Cheng X. et al., 2020), and lncRNA Xist/miR-15a-5p/CUL3signaling pathway (Xu et al., 2019), participated in the SCI, acute kidney injury, and nephropathy procession. Cheng and Wang (2020) identified that lncRNA Xist could act as a ceRNA to sponge miR-212-3p and miR-122-5p to facilitate kidney transplant acute kidney injury progression via regulating the expression of ASF1A, BRWD1, and PFKFB2 using GEO database assay.
In contrast to kidney diseases, the study of lncRNA Xist in cardiovascular diseases is still in its infancy. Chen G. et al. (2018) suggested that lncRNA Xist, which is disrupted by aberrant expression of PFOS (Perfluorooctane sulfonate) in prenatal cells, facilitates placental angiogenesis by regulation of miR-429/VEGF-A axis. Similarly, lncRNA Xist (Hu C. et al., 2019), which positively modulates SOX7 (SRY-box 7) expression by sponging miR-485, participated in hypoxia-induced angiogenesis to activate VEGF signaling pathway, ERK1/2, and Akt signaling pathway through regulation miR-485/SCX7 axis. In additional, Stanford Type A Aortic Dissection (TAAD) is one of the most lethal cardiovascular diseases with an extremely high morbidity and mortality rate. Zhang et al. (2020b) have suggested that lncRNA Xist, which positively regulates PTEN expression via its competitive target miR-17, modulates the proliferation and apoptosis of vascular smooth muscle cells to affect Stanford Type A Aortic Dissection. All in all, these findings provide a new orientation for lncRNA Xist in kidney and cardiovascular diseases.
LncRNA Xist in Other Disease and Cells
A growing number of studies exhibited (Agrelo and Wutz, 2010; Shi et al., 2013; Cantone and Fisher, 2017) that lncRNA Xist participated in disease-associated processes, such as pulmonary disease, diabetic nephropathy, dermal diseases, and hereditary diseases, and mediated cellular functions of cells, such as somatic cell, B cells, and embryonic stem (ES) cells. In acute pneumonia, lncRNA Xist was robustly increased in serum of patients with acute-stage pneumonia and LPS (lipopolysaccharide)-induced WI-38 (normal human fibroblast WI-38 cell line) human lung fibroblasts cells, which shows it is involved in the progression of cell inflammatory response (Zhang et al., 2019g). Consequently, knockdown lncRNA Xist, which functions as a ceRNA to positively modulate TLR4 expression by sponging miR-370-3p, remarkably alleviates LPS-induced cell injury through regulating miR-370-3p/TLR4 axis to activate JAK/STAT and NF−κB signaling pathways (Zhang et al., 2019g).
In pulmonary fibrosis, Wang Y.C. et al. (2017) have revealed that lncRNA Xist regulates bleomycin (BLM)-induced extracellular matrix (ECM) and pulmonary fibrosis via modulation of miR-139/β-catenin axis. Primary graft dysfunction (PGD), which is a major cause of fatality post-lung transplantation, is a known acute lung injury (ALI). Li et al. (2020a) found that lncRNA Xist, which positively elevates the expression of IL-12A by acting as a ceRNA of miR-21, induces NET (neutrophil extracellular trap) formation and accelerates PGD after lung transplantation by activating the network of miR-21/IL-12A.
Diarrhea-predominant irritable bowel syndrome (IBS-D) is prevalent and has a high incidence rate in children. Zhang et al. (2020d) have demonstrated that lncRNA Xist, which is highly expressed in visceral hypersensitivity mice with IBS-D, modulates HT (5-hydroxytrytophan)-induced visceral hypersensitivity by epigenetic silencing of the SERT gene in mice with diarrhea-predominant IBS. In addition, Shen X.F. et al. (2020) have suggested that the silencing of lncRNA Xist, which is highly expressed in serum of patients, protects against sepsis-induced acute liver injury via inhibition of BRD4 expression. In diabetic nephropathy, Wang (2020) reported that lncRNA Xist silencing, which positively modulates PSMB8 expression via acting as a sponge for miR-485 in HMCs (human mesangial cells) treated with high glucose, alleviates inflammation and mesangial cell proliferation via interacting with miR-485/PSMB8.
Autoimmune disorders, such as Hashimoto’s thyroiditis, Sjögren’s Syndrome, systemic lupus erythematosus (SLE), and Grave’s disease, where 85–95% of patients are women, exhibited a strong female bias (Syrett et al., 2019, 2020). In recent years, lncRNA Xist (Syrett et al., 2017, 2019, 2020; Zhang et al., 2020c), which serves a vital function in SLE by RNA-seq data, promotes SLE development in NZB/WF1 mice with lupus-like disease. Taken together, these studies provide an important insight into how lncRNA Xist provides a therapeutic opportunity in female-biased autoimmune disorders.
Rett syndrome (RS), which is a debilitating neurological disorder affecting mostly girls, was caused by heterozygous mutations in the gene encoding the methyl-CpG–binding protein MeCP2 on the X chromosome (Sripathy et al., 2017). lncRNA Xist facilitates RS development through regulation of the bone morphogenetic protein (BMP)/TGF-β signaling pathway (Sripathy et al., 2017), and contributes to mouse brain development through reactivating MeCP2 expression (Adrianse et al., 2018).
Acute respiratory distress syndrome (ARDS), which is associated with diffuse alveolar injury and capillary endothelial damage, is a common clinical syndrome with high a mortality rate (Wang et al., 2019d). lncRNA Xist (Wang et al., 2019d), which acts as a ceRNA to negatively upregulate IRF2 (interferon regulatory factor 2) expression to sponge miR-204, significantly decreases the PaO2/FiO2 ratio and aggravates lipopolysaccharide-induced ARDS in mice by regulating the miR-204/IRF2 axis.
In the parthenogenetic development of pigs, silencing lncRNA Xist remarkedly increased the total blastocyst cell number but did not influence the rate of embryo cleavage and blastocyst formation compared with the control group (Chen et al., 2019e). This study suggested that lncRNA Xist may play a role in a new approach for improving the quality of porcine parthenogenetic embryos. lncRNA Xist (Zhang et al., 2019b) facilitates cells development in somatic cells by TALE-based designer transcriptional factor, and regulates embryonic stem cells’ fates (Chelmicki et al., 2014; An et al., 2020). lncRNA Xist promotes hair follicle regeneration in Dermal papilla cells via regulating miR-424/Shh axis to activate hedgehog signaling (Lin B.J. et al., 2020), and regulates HT cell proliferation and invasion in human trophoblast (HT) cells via miR-144/Titin axis by activating the downstream MAPK and MMPs pathway (Yu N.H. et al., 2017). In polycystic ovary syndrome (PCOS), lncRNA Xist is correlated with adverse pregnancy outcomes (Liu et al., 2020c). In addition, these signaling pathways, which include lncRNA Xist/miR-203-3p/ZFPM2 (Niu et al., 2020), lncRNA Xist/let-7c-5p/STAT3 (Wang et al., 2020d), and lncRNA Xist/miR-320/NOD2 (Xu X.H. et al., 2018), have been identified as a ceRNA regulatory network and participated in osteoblast development and ox-LDL (oxidative low-density lipoprotein)-induced endothelial cells injury.
A previous study reported that lncRNA Xist contributed to human skin fibroblasts by serving as a miRNA sponge. However, lncRNA Xist (Guo et al., 2018; Cao and Feng, 2019) regulates these processes containing skin fibroblasts proliferation, migration, and ECM (extracellular matrix) synthesis after thermal injury by sponging miRNAs (miR-29a and 29b-3p) to promote the expression of target genes (LIN28A and COL1A1). Additionally, lncRNA Xist/miR-181a/COL4A1 axis (Tian R. et al., 2020) is involved in the development and progression of keratoconus using transcriptome RNA-seq data assay. All in all, these results demonstrated that lncRNA Xist plays a pivotal function in non-cancer diseases.
Discussion and Perspectives
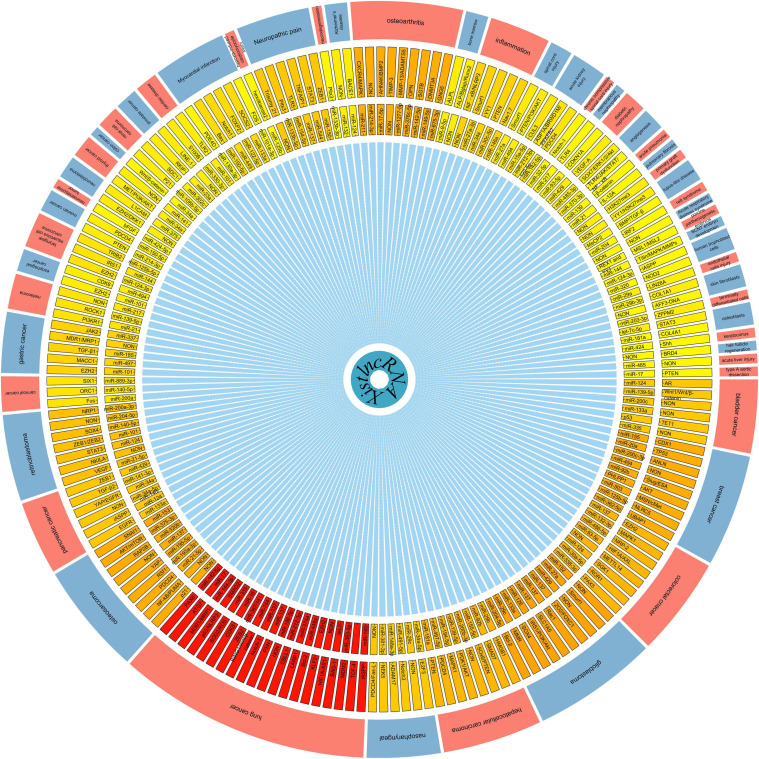
LncRNA Xist, which is conserved among eutherians (human, Rat, mouse, cow, dog, and elephant) but not non-eutherian vertebrates, is an important initiator of the process of XCI in eutherian mammals (Brockdorff et al., 1991; Duret et al., 2006; Galupa et al., 2020). lncRNA Xist is produced by Xist gene and is up-regulated from the Xi chromosome during the XCI process, and recruits protein complexes to reprogram chromosomes [such as H3K27me3 and H2AK119ub trimethylation (Postlmayr et al., 2020)]. In addition to its original XCI functions, numerous studies (Chaligne and Heard, 2014; Dey et al., 2014; Schmitz et al., 2016; Yang Z. et al., 2018; Cheng J.T. et al., 2019; Yan et al., 2019) have also indicated that lncRNA Xist is related to the pathogenic process of multiple diseases by regulating of cell migration, invasion, apoptosis, differentiation, proliferation, and drug resistance. Further investigation of lncRNA, which is considered to function as a miRNA or gene regulator, may aid in addressing disease etiology, such as lung cancer, breast cancer, glioblastoma, osteoarthritis, neuropathic pain, heart disease, and inflammation (Tables 1, 2). By summarizing current knowledge, we noticed that the regulatory network of lncRNA Xist in the majority of biological processions varies considerably. However, lncRNA Xist appears to regulate these processes primarily by interacting with miRNAs to positively facilitate downstream target gene expression (Figure 3). Further studies showed that the regulatory network of lncRNA Xist participated in various signaling pathways, such as TGF-beta signaling pathway, PIK3/AKT signaling pathway, Wnt/β-catenin signaling pathway, FOXO signaling pathway, NF-kB signaling pathway, mTOR signaling pathway, MAPK signaling pathway, Toll-like receptor signaling pathway, JAK-STAT signaling pathway, T cell receptor signaling pathway, and B cell receptor signaling pathway (Tables 1, 2). Although lncRNA Xist taking part in these signaling pathway functions has rarely been demonstrated, there is no reason to believe that the unexplored functions of lncRNA Xist will not be understanded in these ways. These mechanisms of lncRNA Xist action in diseases can indirectly and directly provide recommendations for future research, and more functions of lncRNA Xist can be confirmed.
FIGURE 3.
Overview of the regulatory network of lncRNA Xist involved in mammalian diseases and cells. NON, The downstream target Unclear.
In theory, the genes lncRNA Xist over-expresses and silences are numerous. But over-expression of lncRNA Xist, which is a 15–17 kb RNA polymerase II transcript that is both spliced and polyadenylated (Brockdorff, 2019), is different when using plasmids. By contrast, lncRNA Xist may be inhibited using small molecule inhibitors that block specific binding sites (Matoba et al., 2011). Based on the above reasons, understanding of the function of lncRNA Xist is in its infancy for various diseases and cells. With the developing genome editing technology (Du and Qi, 2016; Chen et al., 2019b; Yi and Li, 2020), CRISPR/Cas9 system is emerging as a powerful tool for sequence-specific control of lncRNA Xist expression in mammalian cells. Recently, numerous studies (Yue and Ogawa, 2018; Colognori et al., 2019; Waśko et al., 2019; Deng et al., 2020) have indicated that CRISPR/Cas9 system is useful for studying lncRNA Xist function and related ceRNA regulatory networks. By combining other future technologies, the function and mechanism of lncRNA Xist will certainly be found. Investigation of lncRNA Xist in virous cells may uncover numerous novel therapeutic approaches for disease treatment in the future. At the same time, it might result in a better understanding of how lncRNA Xist contributes to the XCI and diseases in mammals, potentially opening new avenues for research and therapeutic manipulation of these diseases.
Author Contributions
DZ, WW, and LZ: manuscript design. WW, JM, and XW: literature collection and summary. WW and LZ: drafting of the manuscript. WW, LM, and CL: figure drawing. XQ, WW, DZ, and LZ: revising of the manuscript. All authors have read and approved the final submitted manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank all the members of the research groups of LZ. We apologize to colleagues whose work we were unable to cite or discuses due to space limitations.
Glossary
lncRNAs, Long non-coding RNAs; lncRNA Xist, Long non-coding RNA X-inactive specific transcript; XIC, X chromosome inactivation center; ceRNA, competing endogenous RNA; XCI, X-chromosome inactivation; Xa, active X chromosome; Xi, inactive X chromosome; Ftx, Ftx transcript, Xist regulator; Jpx, Jpx transcript, Xist activator; Tsix, Tsix transcript, Xist antisense RNA; OCT4, also known as Pou5f1; CTCF, CCCTC-binding factor; PRC1 and PRC2, Polycomb repressive complexes (PRCs) -PRC1 and -PRC2; RNA Pol II, RNA polymerase II; hnRNPU (also known as SAF-A), heterogeneous nuclear protein U; ATRX, ATRX chromatin remodeler; hnRNPK, Heterogeneous nuclear ribonucleoprotein K; SHARP, SPEN family transcriptional repressor; HDAC3, histone deacetylase 3; LBR, lamin B receptor; Airn, antisense of IGF2R non-protein coding RNA; Kcnq1ot1, KCNQ1 opposite strand/antisense transcript 1; RBM15, RNA binding motif protein 15; WTAP, WT1 associated protein; U1 snRNP, U1 small nuclear ribonucleoprotein; Rsx, RNA-on-the-silent X; NANOG, Nanog homeobox; SOX2, SRY-box transcription factor 2; PRDM14, PR/SET domain 14; REX1, ZFP42 zinc finger protein; RNF12, RING-H2 finger protein; CdK8, cyclin dependent kinase 8; AR, Androgen Receptor; BCSC, bladder cancer stem cell; PD, Platycodin D; BRCA1, Breast Cancer 1 protein; EMT, epithelial mesenchymal transition; CDX1, caudal-type homeobox 1; TNBC, Triple-negative breast cancer; ESA, epithelium-specific antigen; RELA, NF-κB subunit; BCSCs, Breast cancer stem cells; PHLPP1, PH domain and leucine-rich repeat phosphatase 1; ZEB1, zinc finger E-box binding homeobox 1; EZH2, enhancer of zeste homolog 2; MAPK1, mitogen-activated protein kinase 1; NRP-2, neuropilin-2; HIF-1A, hypoxia inducible factor 1, alpha subunit; METTL14, methyltransferase-like14; YTHDF2, YTHDF proteins 2; SGK1, serum and glucocorticoid-inducible kinase 1; ROR1, receptor-tyrosine-kinase-like orphan receptor 1; Smurf1, smad ubiquitination regulatory factor 1; FOXC1, forkhead box C1; ZO-2, zonula occludens 2; Rac1, Rac family small GTPase 1; ASCT2 and SLC1A5, alanine-, serine-, and cysteine-preferring transporter 2; PI3K, phosphatidylinositol-4,5-bisphosphate 3-kinase; IRS1, insulin receptor substrate 1; SOX4, Y-related high-mobility group box 4; TMZ, Temozolomide; MMR, DNA mismatch repair; HBV, hepatitis B virus; HCV, hepatitis C virus; HMGB1, high-mobility group box-1; PDK1, pyruvate dehydrogenase kinase 1; TNM, tumor-node-metastasis; PDCD4, programmed cell death 4; PTEN, phosphatase and tensin homolog; E2F3, E2F transcription factor 3; ADAM17, a disintegrin and metalloproteinase 17; NEK5, NIMA related kinase 5; TAM, tumor-associated macrophage; TCF-4T, T-cell-specific transcription factor 4; MDM2, mouse double minute clone 2; BAG-1, bcl-2 associated athanogene-1; NSCLC, Non-small cell lung carcinoma; LARP1, La-related protein 1; CBLL1, Casitas B-lineage proto-oncogene like 1; TGF-β1, Transforming growth factor β; ZEB2, zinc finger E-box binding homeobox 2; ATG7, autophagy associated gene 7; PUMA, p53 upregulated modulator of apoptosis; NF-kB, nuclear factor-kappa B; YAP, Hippo/yes-associated protein; EGFR, epidermal growth factor receptor; iASPP, inhibitor of apoptosis-stimulating protein of p53; CDK1, Cyclin-dependent kinase 1; VEGF, vascular endothelial growth factor; STAT3, signal transducer and activator of transcription 3; MACC1, MET transcriptional regulator; MDR1, multidrug resistance gene 1; MRP1, multi-drug resistance protein 1; bFGF, basic fibroblast growth factor; SDC1:Syndecan-1; TGCT, testicular germ cell tumor; CAD, coronary artery disease; MI, myocardial infarction; HF, heart failure; TLR2, toll-like receptor 2; PDE4D, Phosphodiesterase 4D; CB2R, cannabinoid receptor type II; LR5, toll-like receptor 5; NFAIP1, tumor necrosis factor alpha-induced protein 1; CCI, chronic constriction injury; AD, Alzheimer’s disease; OA, Osteoarthritis; PDLSCs, periodontal ligament stem cells; OP, osteoporosis; ALP, alkaline phosphatase; MMP-13, matrix metalloproteinase 13; ADAMTS5, ADAM metallopeptidase with thrombospondin type 1 motif 5; OPN, overexpression of osteopontin; TLRS, Toll−like receptors; CRPS, Complex regional pain syndrome; YY1, Yin-Yang 1; SCI, Spinal cord injury; CCSCI, Compressive Spinal Cord Injury; CDKN1A, cyclin-dependent kinase inhibitor 1A; PFOS, Perfluorooctane sulfonate; SOX7, SRY-box 7; PGD, Primary graft dysfunction; NET, neutrophil extracellular trap; NOD2, Nucleotide-Binding Oligomerization Domain 2; COL1A1, collagen 1 alpha 1.
Footnotes
Funding. This work was supported by grants from the Hunan Provincial Innovation Foundation for Postgraduate (No. 434517123Q), the Natural Science Foundation of Hunan Province (No. 2020JJ5656), and the National Natural Science Foundation of China (31870855).
References
- Adrianse R. L., Smith K., Gatbonton-Schwager T., Sripathy S. P., Lao U., Foss E. J., et al. (2018). Perturbed maintenance of transcriptional repression on the inactive X-chromosome in the mouse brain after Xist deletion. Epigenet. Chromatin. 11 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrelo R., Wutz A. (2010). X inactivation and disease. Semin. Cell Dev. Biol. 21 194–200. [DOI] [PubMed] [Google Scholar]
- An C., Feng G., Zhang J., Cao S., Wang Y., Wang N., et al. (2020). Overcoming autocrine FGF signaling-induced heterogeneity in naive human ESCs enables modeling of random X chromosome inactivation. Cell Stem Cell 27 482.e4–497.e4. [DOI] [PubMed] [Google Scholar]
- Augui S., Nora E. P., Heard E. (2011). Regulation of X-chromosome inactivation by the X-inactivation centre. Nat. Rev. Genet. 12 429–442. 10.1038/nrg2987 [DOI] [PubMed] [Google Scholar]
- Balmant N. V., Reis R. D., Santos M. D., Maschietto M., de Camargo B. (2019). Incidence and mortality of bone cancer among children, adolescents and young adults of Brazil. Clinics 74:e858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar S., Seaton L. R., Weissbein U., Eldar-Geva T., Benvenisty N. (2019). Global characterization of X chromosome inactivation in human pluripotent stem cells. Cell Rep. 27 20.e3–29.e3. [DOI] [PubMed] [Google Scholar]
- Batash R., Asna N., Schaffer P., Francis N., Schaffer M. (2017). Glioblastoma multiforme, diagnosis and treatment; recent literature review. Curr. Med. Chem. 24 3002–3009. [DOI] [PubMed] [Google Scholar]
- Bianchini G., Balko J. M., Mayer I. A., Sanders M. E., Gianni L. (2016). Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 13 674–690. 10.1038/nrclinonc.2016.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bone J. R., Kuroda M. I. (1996). Dosage compensation regulatory proteins and the evolution of sex chromosomes in Drosophila. Genetics 144 705–713. 10.1093/genetics/144.2.705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford C. R., Ferlito A., Devaney K. O., Makitie A. A., Rinaldo A. (2020). Prognostic factors in laryngeal squamous cell carcinoma. Laryngosc. Investig. Otolaryngol. 5 74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J. Clin. 68 394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- Brockdorff N. (2019). Localized accumulation of Xist RNA in X chromosome inactivation. Open Biol. 9 19213–19224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N., Ashworth A., Kay G. F., Cooper P., Smith S., McCabe V. M., et al. (1991). Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature 351 329–331. 10.1038/351329a0 [DOI] [PubMed] [Google Scholar]
- Brown C. J., Hendrich B. D., Rupert J. L., Lafreniere R. G., Xing Y., Lawrence J., et al. (1992). The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71 527–542. 10.1016/0092-8674(92)90520-m [DOI] [PubMed] [Google Scholar]
- Cantone I., Fisher A. G. (2017). Human X chromosome inactivation and reactivation: implications for cell reprogramming and disease. Philos. Trans. R. Soc. B 372:20160358. 10.1098/rstb.2016.0358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H. F., Wahlestedt C., Kapranov P. (2018). Strategies to annotate and characterize long noncoding RNAs: advantages and pitfalls. Trends Genet. 34 704–721. 10.1016/j.tig.2018.06.002 [DOI] [PubMed] [Google Scholar]
- Cao W., Feng Y. P. (2019). LncRNA XIST promotes extracellular matrix synthesis, proliferation and migration by targeting miR-29b-3p/COL1A1 in human skin fibroblasts after thermal injury. Biol. Res. 52 52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassoux N., Lumbroso L., Levy-Gabriel C., Aerts I., Doz F., Desjardins L. (2017). Retinoblastoma: update on current management. Asia Pac. J. Ophthalmol. 6 290–295. [DOI] [PubMed] [Google Scholar]
- Chaligne R., Heard E. (2014). X-chromosome inactivation in development and cancer. FEBS Lett. 588 2514–2522. 10.1016/j.febslet.2014.06.023 [DOI] [PubMed] [Google Scholar]
- Chaligne R., Popova T., Mendoza-Parra M. A., Saleem M. A. M., Gentien D., Ban K., et al. (2015). The inactive X chromosome is epigenetically unstable and transcriptionally labile in breast cancer. Genome Res. 25 488–503. 10.1101/gr.185926.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda K., Mukhopadhyay D. (2020). LncRNA Xist, X- chromosome Instability and Alzheimer’s disease. Curr. Alzheimer Res. 17 499–507. 10.2174/1567205017666200807185624 [DOI] [PubMed] [Google Scholar]
- Chang S. Z., Chen B. H., Wang X. Y., Wu K. Q., Sun Y. Q. (2017). Long non-coding RNA XIST regulates PTEN expression by sponging miR-181a and promotes hepatocellular carcinoma progression. BMC Cancer 17:248. 10.1186/s12885-017-3216-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelmicki T., Dundar F., Turley M. J., Khanam T., Aktas T., Ramirez F., et al. (2014). MOF-associated complexes ensure stem cell identity and Xist repression. eLife 3:e02024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. K., Blanco M., Jackson C., Aznauryan E., Ollikainen N., Surka C., et al. (2016). Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 354 468–472. 10.1126/science.aae0047 [DOI] [PubMed] [Google Scholar]
- Chen D., Chen T., Guo Y., Wang C., Dong L., Lu C. (2020). Platycodin D (PD) regulates LncRNA-XIST/miR-335 axis to slow down bladder cancer progression in vitro and in vivo. Exp. Cell Res. 20:112281. 10.1016/j.yexcr.2020.112281 [DOI] [PubMed] [Google Scholar]
- Chen D. L., Chen L. Z., Lu Y. X., Zhang D. S., Zeng Z. L., Pan Z. Z., et al. (2017). Long noncoding RNA XIST expedites metastasis and modulates epithelial-mesenchymal transition in colorectal cancer. Cell Death Dis. 8:e3011. 10.1038/cddis.2017.421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D. L., Ju H. Q., Lu Y. X., Chen L. Z., Zeng Z. L., Zhang D. S., et al. (2016). Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J. Exp. Clin. Cancer Res. 35 142–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Xu L. L., Huang Y. F., Wang Q., Wang B. H., Yu Z. H., et al. (2018). Prenatal exposure to perfluorooctane sulfonate impairs placental angiogenesis and induces aberrant expression of LncRNA xist. Biomed. Environ. Sci. 31 843–847. [DOI] [PubMed] [Google Scholar]
- Chen H. W., Yang S. D., Shao R. Y. (2019a). Long non-coding XIST raises methylation of TIMP-3 promoter to regulate collagen degradation in osteoarthritic chondrocytes after tibial plateau fracture. Arthrit. Res. Ther. 21 271–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. L. (2016). Linking long noncoding RNA localization and function. Trends Biochem. Sci. 41 761–772. 10.1016/j.tibs.2016.07.003 [DOI] [PubMed] [Google Scholar]
- Chen W., Zhang G. Q., Li J., Zhang X., Huang S. L., Xiang S. L., et al. (2019b). CRISPRlnc: a manually curated database of validated sgRNAs for lncRNAs. Nucleic Acids Res. 47 D63–D68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Zheng R., Baade P. D., Zhang S., Zeng H., Bray F., et al. (2016). Cancer statistics in China, 2015. Cancer J. Clin. 66 115–132. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- Chen X., Xiong D., Ye L., Wang K., Huang L., Mei S., et al. (2019c). Up-regulated lncRNA XIST contributes to progression of cervical cancer via regulating miR-140-5p and ORC1. Cancer Cell Int. 19 45–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Yang L., Ge D. W., Wang W. W., Yin Z. W., Yan J. W., et al. (2019d). Long non-coding RNA XIST promotes osteoporosis through inhibiting bone marrow mesenchymal stem cell differentiation. Exp. Ther. Med. 17 803–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. Y., Zhu Z. W., Yu F. X., Huang J., Jia R. X., Pan J. Z. (2019e). Effect of shRNA-mediated Xist knockdown on the quality of porcine parthenogenetic embryos. Dev. Dyn. 248 140–148. 10.1002/dvdy.24660 [DOI] [PubMed] [Google Scholar]
- Chen Y., Liu X., Chen L., Chen W., Zhang Y., Chen J., et al. (2018). The long noncoding RNA XIST protects cardiomyocyte hypertrophy by targeting miR-330-3p. Biochem. Biophys. Res. Commun. 505 807–815. 10.1016/j.bbrc.2018.09.135 [DOI] [PubMed] [Google Scholar]
- Chen Z., Zhang Y. (2020). Maternal H3K27me3-dependent autosomal and X chromosome imprinting. Nat. Rev. Genet. 21 555–571. 10.1038/s41576-020-0245-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Hu X., Wu Y., Cong L., He X., Lu J., et al. (2019f). Long non-coding RNA XIST promotes the development of esophageal cancer by sponging miR-494 to regulate CDK6 expression. Biomed. Pharmacother. 109 2228–2236. 10.1016/j.biopha.2018.11.049 [DOI] [PubMed] [Google Scholar]
- Cheng J. T., Wang L. Z., Wang H., Tang F. R., Cai W. Q., Sethi G., et al. (2019). Insights into biological role of LncRNAs in epithelial-mesenchymal transition. Cells Basel 8 1178–1203. 10.3390/cells8101178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q., Wang L. (2020). LncRNA XIST serves as a ceRNA to regulate the expression of ASF1A, BRWD1M, and PFKFB2 in kidney transplant acute kidney injury via sponging hsa-miR-212-3p and hsa-miR-122-5p. Cell Cycle 19 290–299. 10.1080/15384101.2019.1707454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q., Xu X. Y., Jiang H., Xu L. E., Li Q. (2018). Knockdown of long non-coding RNA XIST suppresses nasopharyngeal carcinoma progression by activating miR-491-5p. J. Cell. Biochem. 119 3936–3944. 10.1002/jcb.26535 [DOI] [PubMed] [Google Scholar]
- Cheng X., Xu J., Yu Z., Xu J., Long H. (2020). LncRNA xist contributes to endogenous neurological repair after chronic compressive spinal cord injury by promoting angiogenesis through the miR-32-5p/Notch-1 axis. J. Cell Sci. 8 744–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Chang Q., Zheng B., Xu J., Li H., Wang R. (2019). LncRNA XIST promotes the epithelial to mesenchymal transition of retinoblastoma via sponging miR-101. Eur. J. Pharmacol. 843 210–216. 10.1016/j.ejphar.2018.11.028 [DOI] [PubMed] [Google Scholar]
- Cheng Z. H., Li Z. S. N., Ma K., Li X. Y., Tian N., Duan J. Y., et al. (2017). Long Non-coding RNA XIST Promotes Glioma Tumorigenicity and Angiogenesis by Acting as a Molecular Sponge of miR-429. J. Cancer 8 4106–4116. 10.7150/jca.21024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z. H., Luo C., Guo Z. L. (2020). LncRNA-XIST/microRNA-126 sponge mediates cell proliferation and glucose metabolism through the IRS1/PI3K/Akt pathway in glioma. J. Cell. Biochem. 121 2170–2183. 10.1002/jcb.29440 [DOI] [PubMed] [Google Scholar]
- Chigi Y., Sasaki H., Sado T. (2017). The 5 ‘ region of Xist RNA has the potential to associate with chromatin through the A-repeat. RNA 23 1894–1901. 10.1261/rna.062158.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C., Zhang Q. F. C., da Rocha S. T., Flynn R. A., Bharadwaj M., Calabrese J. M., et al. (2015). Systematic discovery of Xist RNA binding proteins. Cell 161 404–416. 10.1016/j.cell.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua M. L. K., Wee J. T. S., Hui E. P., Chan A. T. C. (2016). Nasopharyngeal carcinoma. Lancet 387 1012–1024. [DOI] [PubMed] [Google Scholar]
- Colognori D., Sunwoo H., Kriz A. J., Wang C. Y., Lee J. T. (2019). Xist deletional analysis reveals an interdependency between xist RNA and polycomb complexes for spreading along the inactive X. Mol. Cell 74 101–117. 10.1016/j.molcel.2019.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colognori D., Sunwoo H., Wang D., Wang C. Y., Lee J. T. (2020). Xist Repeats A and B account for two distinct phases of X inactivation establishment. Dev. Cell 54 21.e5–32.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colpaert R. M. W., Calore M. (2019). MicroRNAs in cardiac diseases. Cells Basel 8 737–759. 10.3390/cells8070737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connerty P., Lock R. B., de Bock C. E. (2020). Long non-coding RNAs: major regulators of cell stress in cancer. Front. Oncol. 10:285. 10.3389/fonc.2020.00285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C. L., Li Y. N., Cui X. Y., Wu X. (2020). lncRNA XIST promotes the progression of laryngeal squamous cell carcinoma by sponging miR144 to regulate IRS1 expression. Oncol. Rep. 43 525–535. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Czerminski J. T., Lawrence J. B. (2020). Silencing trisomy 21 with XIST in neural stem cells promotes neuronal differentiation. Dev. Cell 52 294–308. 10.1016/j.devcel.2019.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha S. T., Heard E. (2017). Novel players in X inactivation: insights into Xist-mediated gene silencing and chromosome conformation. Nat. Struct. Mol. Biol. 24 197–204. 10.1038/nsmb.3370 [DOI] [PubMed] [Google Scholar]
- Debrand E., Heard E., Avner P. (1998). Cloning and localization of the murine Xpct gene: evidence for complex rearrangements during the evolution of the region around the Xist gene. Genomics 48 296–303. 10.1006/geno.1997.5173 [DOI] [PubMed] [Google Scholar]
- Deng C., Hu X., Wu K., Tan J., Yang C. (2019). Long non-coding RNA XIST predicting advanced clinical parameters in cancer: a Meta-Analysis and case series study in a single institution. Oncol. Lett. 18 2192–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M., Liu Z., Chen B., Cai Y., Wan Y., Wang F. (2020). Locus-specific regulation of xist expression using the CRISPR-Cas9-based system. DNA Cell Biol. 39 572–578. 10.1089/dna.2019.4945 [DOI] [PubMed] [Google Scholar]
- Deng X., Berletch J. B., Nguyen D. K., Disteche C. M. (2014). X chromosome regulation: diverse patterns in development, tissues and disease. Nat. Rev. Genet. 15 367–378. 10.1038/nrg3687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22 1775–1789. 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux Y., Zangrando J., Schroen B., Creemers E. E., Pedrazzini T., Chang C. P., et al. (2015). Long noncoding RNAs in cardiac development and ageing. Nat. Rev. Cardiol. 12 415–425. 10.1038/nrcardio.2015.55 [DOI] [PubMed] [Google Scholar]
- Dey B. K., Mueller A. C., Dutta A. (2014). Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription 5:e944014. 10.4161/21541272.2014.944014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinescu S., Ignat S., Lazar A. D., Constantin C., Neagu M., Costache M. (2019). Epitranscriptomic signatures in lncRNAs and their possible roles in cancer. Genes 10 52–79. 10.3390/genes10010052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disteche C. M. (2012). Dosage compensation of the sex chromosomes. Annu. Rev. Genet. 46 537–560. 10.1146/annurev-genet-110711-155454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disteche C. M. (2016). Dosage compensation of the sex chromosomes and autosomes. Semin. Cell Dev. Biol. 56 9–18. 10.1016/j.semcdb.2016.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Wan G., Peng G., Yan P., Qian C., Li F. (2020). Long non-coding RNA XIST regulates hyperglycemia-associated apoptosis and migration in human retinal pigment epithelial cells. Biomed. Pharmacother. 125:109959. 10.1016/j.biopha.2020.109959 [DOI] [PubMed] [Google Scholar]
- Donohoe M. E., Silva S. S., Pinter S. F., Xu N., Lee J. T. (2009). The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature 460 128–U147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dossin F., Pinheiro I., Zylicz J. J., Roensch J., Collombet S., Saux A. Le, et al. (2020). SPEN integrates transcriptional and epigenetic control of X-inactivation. Nature 578 455–460. 10.1038/s41586-020-1974-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du D., Qi L. S. (2016). CRISPR technology for genome activation and repression in mammalian cells. Cold Spring Harb. Protoc. 2016:rot090175. [DOI] [PubMed] [Google Scholar]
- Du P., Zhao H. T., Peng R. J., Liu Q., Yuan J., Peng G., et al. (2017). LncRNA-XIST interacts with miR-29c to modulate the chemoresistance of glioma cell to TMZ through DNA mismatch repair pathway. Biosci. Rep. 37:BSR20170696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Gao G. Q., Li J. X., Suo S., Liu S., Sun Y., et al. (2020). Silencing of long noncoding RNA XIST attenuated Alzheimer’s disease-related BACE1 alteration through miR-124. Cell Biol. Int. 44 630–636. 10.1002/cbin.11263 [DOI] [PubMed] [Google Scholar]
- Du Y., Weng X. D., Wang L., Liu X. H., Zhu H. C., Guo J., et al. (2017). LncRNA XIST acts as a tumor suppressor in prostate cancer through sponging miR-23a to modulate RKIP expression. Oncotarget 8 94358–94370. 10.18632/oncotarget.21719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L., Chureau C., Samain S., Weissenbach J., Avner P. (2006). The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 312 1653–1655. 10.1126/science.1126316 [DOI] [PubMed] [Google Scholar]
- Eberlin K. R. (2019). Invited commentary targeted muscle reinnervation: a significant advance in the prevention and treatment of post-amputation neuropathic pain. J. Am. Coll. Surgeons 228 226–227. 10.1016/j.jamcollsurg.2018.12.008 [DOI] [PubMed] [Google Scholar]
- Fan J. L., Zhu T. T., Xue Z. Y., Ren W. Q., Guo J. Q., Zhao H. Y., et al. (2020). lncRNA-XIST protects the hypoxia-induced cardiomyocyte injury through regulating the miR-125b-hexokianse 2 axis. In Vitro Cell Dev. An. 56 349–357. 10.1007/s11626-020-00459-0 [DOI] [PubMed] [Google Scholar]
- Fang J., Sun C. C., Gong C. (2016). Long noncoding RNA XIST acts as an oncogene in non-small cell lung cancer by epigenetically repressing KLF2 expression. Biochem. Biophys. Res. Commun. 478 811–817. 10.1016/j.bbrc.2016.08.030 [DOI] [PubMed] [Google Scholar]
- Feng Y. M., Wan P. B., Yin L. L. (2020). Long noncoding RNA X-inactive specific transcript (XIST) promotes osteogenic differentiation of periodontal ligament stem cells by sponging MicroRNA-214-3p. Med. Sci. Monitor 26 e918931–e918932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F., Alekseyenko A. A., Park P. J., Kuroda M. I. (2014). Transcriptional control of a whole chromosome: emerging models for dosage compensation. Nat. Struct. Mol. Biol. 21 118–125. 10.1038/nsmb.2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finestra T. R., Gribnau J. (2017). X chromosome inactivation: silencing, topology and reactivation. Curr. Opin. Cell Biol. 46 54–61. 10.1016/j.ceb.2017.01.007 [DOI] [PubMed] [Google Scholar]
- Fornasari D. (2017). Pharmacotherapy for neuropathic pain: a review. Pain Ther. 6 25–33. 10.1007/s40122-017-0091-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. D. (2014). Non-coding RNA: a new frontier in regulatory biology. Natl. Sci. Rev. 1 190–204. 10.1093/nsr/nwu008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlan G., Rougeulle C. (2016). Function and evolution of the long noncoding RNA circuitry orchestrating X-chromosome inactivation in mammals. Wires RNA 7 702–722. 10.1002/wrna.1359 [DOI] [PubMed] [Google Scholar]
- Galupa R., Nora E. P., Worsley-Hunt R., Picard C., Gard C., van Bemmel J. G., et al. (2020). A conserved noncoding locus regulates random monoallelic xist expression across a topological boundary. Mol. Cell 77 352–367. 10.1016/j.molcel.2019.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Gao J., Chen L., Ren Y., Ma J. (2019). Targeting XIST induced apoptosis of human osteosarcoma cells by activation of NF-kB/PUMA signal. Bioengineered 10 261–270. 10.1080/21655979.2019.1631104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayen S., Maclary E., Buttigieg E., Hinten M., Kalantry S. (2015). A primary role for the tsix lncRNA in maintaining random X-chromosome inactivation. Cell Rep. 11 1251–1265. 10.1016/j.celrep.2015.04.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel A. V., Heard E. (2014). Noncoding RNAs and epigenetic mechanisms during X-chromosome inactivation. Annu. Rev. Cell Dev. Biol. 30 561–580. 10.1146/annurev-cellbio-101512-122415 [DOI] [PubMed] [Google Scholar]
- Ghaderian S. M. H., Alipour Parsa S., Ghaedi H., Jafari H., Sun P., Wu Y. (2020). miR-142-5p protects against osteoarthritis through competing with lncRNA XIST. Arch. Physiol. Biochem. 22:e3158. [DOI] [PubMed] [Google Scholar]
- Giorgetti L., Lajoie B. R., Carter A. C., Attia M., Zhan Y., Xu J., et al. (2016). Structural organization of the inactive X chromosome in the mouse. Nature 535 575–579. 10.1038/nature18589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontan C., Jonkers I., Gribnau J. (2011). “Long noncoding RNAs and X chromosome inactivation,” in Long Non-Coding RNAs, ed. Ugarkovic D. (Berlin: Springer; ), 43–64. 10.1007/978-3-642-16502-3_3 [DOI] [PubMed] [Google Scholar]
- Gontan C., Achame E. M., Demmers J., Barakat T. S., Rentmeester E., van I. W., et al. (2012). RNF12 initiates X-chromosome inactivation by targeting REX1 for degradation. Nature 485 386–390. 10.1038/nature11070 [DOI] [PubMed] [Google Scholar]
- Grant J., Mahadevaiah S. K., Khil P., Sangrithi M. N., Royo H., Duckworth J., et al. (2012). Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature 487 254–258. 10.1038/nature11171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco S., Salgado Somoza A., Devaux Y., Martelli F. (2018). Long noncoding RNAs and cardiac disease. Antioxid. Redox Signal. 29 880–901. [DOI] [PubMed] [Google Scholar]
- Gu S. X., Xie R., Liu X. D., Shou J. J., Gu W. T., Che X. M. (2017). Long Coding RNA XIST Contributes to Neuronal Apoptosis through the Downregulation of AKT phosphorylation and is negatively regulated by miR-494 in rat spinal cord injury. Int. J. Mol. Sci. 18 732–749. 10.3390/ijms18040732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Callaway J. B., Ting J. P. (2015). Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat. Med. 21 677–687. 10.1038/nm.3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Huang X., Liang P. F., Zhang P. H., Zhang M. H., Ren L. C., et al. (2018). Role of XIST/miR-29a/LIN28A pathway in denatured dermis and human skin fibroblasts (HSFs) after thermal injury. J. Cell. Biochem. 119 1463–1474. 10.1002/jcb.26307 [DOI] [PubMed] [Google Scholar]
- Hai B. (2020). LncRNA XIST promotes growth of human chordoma cells by regulating miR-124-3p/iASPP pathway. J. Cell. Physiol. 13 4755–4765. 10.2147/ott.s252195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Shen X. (2020). Long noncoding RNAs in osteosarcoma via various signaling pathways. J. Clin. Lab. Anal. 34:e23317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q., Li L., Liang H., Li Y., Xie J., Wang Z. (2017). Downregulation of lncRNA X inactive specific transcript (XIST) suppresses cell proliferation and enhances radiosensitivity by upregulating mir-29c in nasopharyngeal carcinoma cells. Med. Sci. Monit. 23 4798–4807. 10.12659/msm.905370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper M. I., Fosten M., Monk M. (1982). Preferential paternal X inactivation in extraembryonic tissues of early mouse embryos. J. Embryol. Exp. Morphol. 67 127–135. 10.1242/dev.67.1.127 [DOI] [PubMed] [Google Scholar]
- Hasegawa Y., Brockdorff N., Kawano S., Tsutui K., Tsutui K., Nakagawa S. (2010). The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev. Cell 19 469–476. 10.1016/j.devcel.2010.08.006 [DOI] [PubMed] [Google Scholar]
- He X. L., Yang L., Huang R. Q., Lin L. J., Shen Y. J., Cheng L. M., et al. (2020). Activation of CB2R with AM1241 ameliorates neurodegeneration via the Xist/miR-133b-3p/Pitx3 axis. J. Cell. Physiol. 235 6032–6042. 10.1002/jcp.29530 [DOI] [PubMed] [Google Scholar]
- Hu B., Shi G., Li Q., Li W., Zhou H. (2019). Long noncoding RNA XIST participates in bladder cancer by downregulating p53 via binding to TET1. J. Cell. Biochem. 120 6330–6338. 10.1002/jcb.27920 [DOI] [PubMed] [Google Scholar]
- Hu C., Bai X., Liu C., Hu Z. (2019). Long noncoding RNA XIST participates hypoxia-induced angiogenesis in human brain microvascular endothelial cells through regulating miR-485/SOX7 axis. Am. J. Transl. Res. 11 6487–6497. [PMC free article] [PubMed] [Google Scholar]
- Hu C., Liu S., Han M., Wang Y., Xu C. (2018). Knockdown of lncRNA XIST inhibits retinoblastoma progression by modulating the miR-124/STAT3 axis. Biomed. Pharmacother. 107 547–554. 10.1016/j.biopha.2018.08.020 [DOI] [PubMed] [Google Scholar]
- Hu W. N., Duan Z. Y., Wang Q., Zhou D. H. (2019). The suppression of ox-LDL-induced inflammatory response and apoptosis of HUVEC by lncRNA XIAT knockdown via regulating miR-30c-5p/PTEN axis. Eur. Rev. Med. Pharmacol. 23 7628–7638. [DOI] [PubMed] [Google Scholar]
- Hu Y., Deng C., Zhang H., Zhang J., Peng B., Hu C. (2017). Long non-coding RNA XIST promotes cell growth and metastasis through regulating miR-139-5p mediated Wnt/beta-catenin signaling pathway in bladder cancer. Oncotarget 8 94554–94568. 10.18632/oncotarget.21791 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Huang Y. S., Chang C. C., Lee S. S., Jou Y. S., Shih H. M. (2016). Xist reduction in breast cancer upregulates AKT phosphorylation via HDAC3-mediated repression of PHLPP1 expression. Oncotarget 7 43256–43266. 10.18632/oncotarget.9673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J. Y., Oh J. N., Park C. H., Lee D. K., Lee C. K. (2015). Dosage compensation of X-chromosome inactivation center-linked genes in porcine preimplantation embryos: non-chromosome-wide initiation of X-chromosome inactivation in blastocysts. Mech. Dev. 138 246–255. 10.1016/j.mod.2015.10.005 [DOI] [PubMed] [Google Scholar]
- Jegu T., Aeby E., Lee J. T. (2017). The X chromosome in space. Nat. Rev. Genet. 18 377–389. [DOI] [PubMed] [Google Scholar]
- Jeon Y., Lee J. T. (2011). YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146 119–133. 10.1016/j.cell.2011.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H. J., Zhang H. Z., Hu X. G., Li W. B. (2018). Knockdown of long non-coding RNA XIST inhibits cell viability and invasion by regulating miR-137/PXN axis in non-small cell lung cancer. Int. J. Biol. Macromol. 111 623–631. 10.1016/j.ijbiomac.2018.01.022 [DOI] [PubMed] [Google Scholar]
- Jiang J., Jing Y., Cost G. J., Chiang J. C., Kolpa H. J., Cotton A. M., et al. (2013). Translating dosage compensation to trisomy 21. Nature 500 296–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q., Xing W., Cheng J., Yu Y. (2020). Knockdown of lncRNA XIST suppresses cell tumorigenicity in human non-small cell lung cancer by regulating miR-142-5p/PAX6 axis. OncoTargets Ther. 13 4919–4929. 10.2147/ott.s238808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Lv Y. L., Wang S. Y. (2018). Prediction of non-coding RNAs as drug targets. Adv. Exp. Med. Biol. 1094 109–115. 10.1007/978-981-13-0719-5_11 [DOI] [PubMed] [Google Scholar]
- Jin H., Du X. J., Zhao Y., Xia D. L. (2018). XIST/miR-544 axis induces neuropathic pain by activating STAT3 in a rat model. J. Cell. Physiol. 233 5847–5855. 10.1002/jcp.26376 [DOI] [PubMed] [Google Scholar]
- Jin L. W., Pan M., Ye H. Y., Zheng Y., Chen Y., Huang W. W., et al. (2019). Down-regulation of the long non-coding RNA XIST ameliorates podocyte apoptosis in membranous nephropathy via the miR-217-TLR4 pathway. Exp. Physiol. 104 220–230. [DOI] [PubMed] [Google Scholar]
- Kawakami T., Zhang C., Taniguchi T., Kim C. J., Okada Y., Sugihara H., et al. (2004). Characterization of loss-of-inactive X in Klinefelter syndrome and female-derived cancer cells. Oncogene 23 6163–6169. 10.1038/sj.onc.1207808 [DOI] [PubMed] [Google Scholar]
- Kay G. F. (1998). Xist and X chromosome inactivation. Mol. Cell. Endocrinol. 140 71–76. [DOI] [PubMed] [Google Scholar]
- Khamlichi A. A., Feil R. (2018). Parallels between mammalian mechanisms of monoallelic gene expression. Trends Genet. 34 954–971. 10.1016/j.tig.2018.08.005 [DOI] [PubMed] [Google Scholar]
- Kim M. M., Umemura Y., Leung D. (2018). Bevacizumab and glioblastoma: past, present, and future directions. Cancer J. 24 180–186. 10.1097/ppo.0000000000000326 [DOI] [PubMed] [Google Scholar]
- Kolpa H. J., Fackelmayer F. O., Lawrence J. B. (2016). SAF-A requirement in anchoring XIST RNA to chromatin varies in transformed and primary cells. Dev. Cell 39 9–10. 10.1016/j.devcel.2016.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q. L., Zhang S. Q., Liang C. Q., Zhang Y., Kong Q. C., Chen S. X., et al. (2018). LncRNA XIST functions as a molecular sponge of miR-194-5p to regulate MAPK1 expression in hepatocellular carcinoma cell. J. Cell. Biochem. 119 4458–4468. 10.1002/jcb.26540 [DOI] [PubMed] [Google Scholar]
- Kraus V. B. (2014). Osteoarthritis: the zinc link. Nature 507 441–442. [DOI] [PubMed] [Google Scholar]
- Kung J. T., Kesner B., An J. Y., Ahn J. Y., Cifuentes-Rojas C., Colognori D., et al. (2015). Locus-specific targeting to the X chromosome revealed by the RNA interactome of CTCF. Mol. Cell 57 361–375. 10.1016/j.molcel.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung J. T. Y., Colognori D., Lee J. T. (2013). Long noncoding RNAs: past, present, and future. Genetics 193 651–669. 10.1534/genetics.112.146704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane C. A., Hardy J., Schott J. M. (2018). Alzheimer’s disease. Eur. J. Neurol. 25 59–70. [DOI] [PubMed] [Google Scholar]
- Laner T., Schulz W. A., Engers R., Muller M., Florl A. R. (2005). Hypomethylation of the XIST gene promoter in prostate cancer. Oncol. Res. 15 257–264. 10.3727/096504005776404607 [DOI] [PubMed] [Google Scholar]
- Larsson J., Meller V. H. (2006). Dosage compensation, the origin and the afterlife of sex chromosomes. Chromosome Res. 14 417–431. 10.1007/s10577-006-1064-3 [DOI] [PubMed] [Google Scholar]
- Lee H. J., Gopalappa R., Sunwoo H., Choi S. W., Ramakrishna S., Lee J. T., et al. (2019). En bloc and segmental deletions of human XIST reveal X chromosome inactivation-involving RNA elements. Nucleic Acids Res. 47 3875–3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. T., Bartolomei M. S., Inactivation X. - (2013). Imprinting, and long noncoding RNAs in health and disease. Cell 152 1308–1323. 10.1016/j.cell.2013.02.016 [DOI] [PubMed] [Google Scholar]
- Levey A. S., James M. T. (2017). Acute kidney injury. Ann. Intern. Med. 167 66–80. [DOI] [PubMed] [Google Scholar]
- Li C., Wan L., Liu Z., Xu G., Wang S., Su Z., et al. (2018). Long non-coding RNA XIST promotes TGF-beta-induced epithelial-mesenchymal transition by regulating miR-367/141-ZEB2 axis in non-small-cell lung cancer. Cancer Lett. 418 185–195. 10.1016/j.canlet.2018.01.036 [DOI] [PubMed] [Google Scholar]
- Li G. L., Wu Y. X., Li Y. M., Li J. (2017). High expression of long non-coding RNA XIST in osteosarcoma is associated with cell proliferation and poor prognosis. Eur. Rev. Med. Pharmacol. 21 2829–2834. [PubMed] [Google Scholar]
- Li H., Cui J., Xu B., He S., Yang H., Liu L. (2019). Long non-coding RNA XIST serves an oncogenic role in osteosarcoma by sponging miR-137. Exp Ther. Med. 17 730–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., He D. (2019). Long noncoding RNA Xist predicts the presence of lymph node metastases in human oesophageal squamous cell carcinoma. Br. J. Biomed. Sci. 76 147–149. 10.1080/09674845.2019.1594489 [DOI] [PubMed] [Google Scholar]
- Li J. W., Wei L., Han Z. J., Chen Z., Zhang Q. (2020a). Long non-coding RNA X-inactive specific transcript silencing ameliorates primary graft dysfunction following lung transplantation through microRNA-21-dependent mechanism. EBioMedicine 52 102600–102613. 10.1016/j.ebiom.2019.102600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Lv G. H., Wang B., Kuang L. (2018). The role of lncRNA XIST/miR-211 axis in modulating the proliferation and apoptosis of osteoarthritis chondrocytes through CXCR4 and MAPK signaling. Biochem. Biophys. Res. Commun. 503 2555–2562. 10.1016/j.bbrc.2018.07.015 [DOI] [PubMed] [Google Scholar]
- Li L., Lv G., Wang B., Kuang L. (2020b). XIST/miR-376c-5p/OPN axis modulates the influence of proinflammatory M1 macrophages on osteoarthritis chondrocyte apoptosis. J. Cell. Physiol. 235 281–293. 10.1002/jcp.28968 [DOI] [PubMed] [Google Scholar]
- Li M., Pan M., You C. Z., Zhao F. S., Wu D., Guo M., et al. (2020c). MiR-7 reduces the BCSC subset by inhibiting XIST to modulate the miR-92b/Slug/ESA axis and inhibit tumor growth. Breast Cancer Res. 22 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Hou L., Yin L., Zhao S. (2020d). LncRNA XIST interacts with miR-454 to inhibit cells proliferation, epithelial mesenchymal transition and induces apoptosis in triple-negative breast cancer. J. Biosciences 45 1–11. [PubMed] [Google Scholar]
- Li X., Jia Y., Zong Y., Zhang Y., Hou D., Xu J., et al. (2020e). The lncRNA XIST promotes the progression of breast cancer by sponging miR-125b-5p to modulate NLRC5. J. Gene Med. 12 3501–3511. [PMC free article] [PubMed] [Google Scholar]
- Li Y., Liang Y., Zhu Y., Zhang Y., Bei Y. (2018). Noncoding RNAs in cardiac hypertrophy. J. Cardiovasc. Transl. Res. 11 439–449. 10.1007/s12265-018-9797-x [DOI] [PubMed] [Google Scholar]
- Li Y. D., Zhang Q., Tang X. Q. (2019). Long non-coding RNA XIST contributes into drug resistance of gastric cancer cell. Minerva Med. 110 270–273. [DOI] [PubMed] [Google Scholar]
- Li Z. Q., Zhang Y. P., Ding N., Zhao Y. D., Ye Z. K., Shen L., et al. (2019). Inhibition of lncRNA XIST improves myocardial I/R injury by targeting miR-133a through inhibition of autophagy and regulation of SOCS2. Mol. Ther. Nucl. Acids 18 764–773. 10.1016/j.omtn.2019.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S., Gong X., Zhang G., Huang G., Lu Y., Li Y. (2017). The lncRNA XIST interacts with miR-140/miR-124/iASPP axis to promote pancreatic carcinoma growth. Oncotarget 8 113701–113718. 10.18632/oncotarget.22555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X. Y., Tang D. Z., Yang H. S., Chen Y., Chen D. Y., Jia L. S., et al. (2019). Long non-coding RNA XIST may influence cervical ossification of the posterior longitudinal ligament through regulation of miR-17-5P/AHNAK/BMP2 signaling pathway. Calcif. Tissue Int. 105 670–680. 10.1007/s00223-019-00608-y [DOI] [PubMed] [Google Scholar]
- Librizzi M., Chiarelli R., Bosco L., Sansook S., Gascon J. M., Spencer J., et al. (2015). The histone deacetylase inhibitor JAHA down-regulates pERK and global DNA methylation in MDA-MB231 breast cancer cells. Materials 8 7041–7047. 10.3390/ma8105358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B. J., Zhu J. Y., Ye J., Lu S. D., Liao M. D., Meng X. C., et al. (2020). LncRNA-XIST promotes dermal papilla induced hair follicle regeneration by targeting miR-424 to activate hedgehog signaling. Cell. Signal. 72 109623–109633. 10.1016/j.cellsig.2020.109623 [DOI] [PubMed] [Google Scholar]
- Lin B., Xu J., Wang F., Wang J., Zhao H., Feng D. (2020). LncRNA XIST promotes myocardial infarction by regulating FOS through targeting miR-101a-3p. J. Cell. Mol. Med. 12 7232–7247. 10.18632/aging.103072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X. Q., Huang Z. M., Chen X., Wu F., Wu W. (2018). XIST induced by JPX suppresses hepatocellular carcinoma by sponging miR-155-5p. Yonsei Med. J. 59 816–826. 10.3349/ymj.2018.59.7.816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A., Liu L., Lu H. (2019a). LncRNA XIST facilitates proliferation and epithelial-mesenchymal transition of colorectal cancer cells through targeting miR-486-5p and promoting neuropilin-2. J. Cell. Physiol. 234 13747–13761. 10.1002/jcp.28054 [DOI] [PubMed] [Google Scholar]
- Liu B., Luo C., Lin H., Ji X., Zhang E., Li X. (2020a). Long noncoding RNA XIST acts as a ceRNA of miR-362-5p to suppress breast cancer progression. Cancer Biother. Radiopharm. [Epub ahead of print]. 10.7150/jca.45676 [DOI] [PubMed] [Google Scholar]
- Liu C., Lu Z., Liu H., Zhuang S., Guo P. (2020b). LncRNA XIST promotes the progression of laryngeal squamous cell carcinoma via sponging miR-125b-5p to modulate TRIB2. Bioscience Rep. 40:BSR20193172. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu H., Deng H. Y., Zhao Y. J., Li C., Liang Y. (2018c). LncRNA XIST/miR-34a axis modulates the cell proliferation and tumor growth of thyroid cancer through MET-PI3K-AKT signaling. J. Exp. Clin. Cancer Res. 37 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. L., Yao L., Zhang M. Y., Jiang J., Yang M. P., Wang Y. (2019b). Downregulation of LncRNA-XIST inhibited development of non-small cell lung cancer by activating miR-335/SOD2/ROS signal pathway mediated pyroptotic cell death. Aging Us 11 7830–7846. 10.18632/aging.102291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. L., Zhang W. Q., Zhao M., Huang M. Y. (2019c). Upregulation of long noncoding RNA XIST is associated with poor prognosis in human cancers. J. Cell. Physiol. 234 6594–6600. 10.1002/jcp.27400 [DOI] [PubMed] [Google Scholar]
- Liu M., Zhu H., Li Y., Zhuang J., Cao T., Wang Y. (2020c). Expression of serum lncRNA-Xist in patients with polycystic ovary syndrome and its relationship with pregnancy outcome. Taiwan J. Obstet. Gynecol. 59 372–376. 10.1016/j.tjog.2020.03.006 [DOI] [PubMed] [Google Scholar]
- Liu W. G., Xu Q. (2019). Long non-coding RNA XIST promotes hepatocellular carcinoma progression by sponging miR-200b-3p. Eur. Rev. Med. Pharmacol. 23 9857–9862. [DOI] [PubMed] [Google Scholar]
- Liu X., Cui L., Hua D. (2018a). Long noncoding RNA XIST regulates miR-137-EZH2 axis to promote tumor metastasis in colorectal cancer. Oncol. Res. 27 99–106. 10.3727/096504018x15195193936573 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu X., Ming X., Jing W., Luo P., Li N., Zhu M., et al. (2018b). Long non-coding RNA XIST predicts worse prognosis in digestive system tumors: a systemic review and meta-analysis. Bioscience Rep. 38:BSR20180169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Xie S., Zhang J., Kang Y. (2020d). Long noncoding RNA XIST contributes to cervical cancer development through targeting miR-889-3p/SIX1 axis. Cancer Biother. Radiopharm. [Epub ahead f print]. [DOI] [PubMed] [Google Scholar]
- Liu Y., Liu K., Tang C., Shi Z., Jing K., Zheng J. (2020e). Long non-coding RNA XIST contributes to osteoarthritis progression via miR-149-5p/DNMT3A axis. Biomed. Pharmacother. 128 110349–110359. 10.1016/j.biopha.2020.110349 [DOI] [PubMed] [Google Scholar]
- Lobo J., Nunes S. P., Gillis A. J. M., Barros-Silva D., Miranda-Goncalves V., van den Berg A., et al. (2019). XIST-promoter demethylation as tissue biomarker for testicular germ cell tumors and spermatogenesis quality. Cancers 11 1385–1400. 10.3390/cancers11091385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loda A., Heard E. (2019). Xist RNA in action: past, present, and future. PLoS Genet. 15:e1008333. 10.1371/journal.pgen.1008333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzen J. M., Thum T. (2016). Long noncoding RNAs in kidney and cardiovascular diseases. Nat. Rev. Nephrol. 12 360–373. 10.1038/nrneph.2016.51 [DOI] [PubMed] [Google Scholar]
- Lu Z., Carter A. C., Chang H. Y. (2017). Mechanistic insights in X-chromosome inactivation. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 372:20160356. 10.1098/rstb.2016.0356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C. X., Quan Z. P., Zhong B., Zhang M., Zhou B., Wang S. B., et al. (2020). lncRNA XIST promotes glioma proliferation and metastasis through miR-133a/SOX4. Exp. Ther. Med. 19 1641–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Luo P., Zhang Y. (2020). Identification of differentially expressed long non-coding RNAs associated with dilated cardiomyopathy using integrated bioinformatics approaches. J. Clin. Lab. Anal. 14 181–186. 10.5582/ddt.2020.01010 [DOI] [PubMed] [Google Scholar]
- Lv G. Y., Miao J., Zhang X. L. (2018). Long noncoding RNA XIST promotes osteosarcoma progression by targeting ras-related protein RAP2B via miR-320b. Oncol. Res. 26 837–846. 10.3727/096504017x14920318811721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu X., Ma Y., Wu F., Wang L., Wang L. (2019). LncRNA NKILA Inhibits Retinoblastoma by Downregulating lncRNA XIST. Curr. Eye Res. 44 975–979. 10.1080/02713683.2019.1606253 [DOI] [PubMed] [Google Scholar]
- Ma L., Bajic V. B., Zhang Z. (2013). On the classification of long non-coding RNAs. RNA Biol. 10 925–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Zhou Y. J., Luo X. J., Gao H., Deng X. B., Jiang Y. J. (2017). Long non-coding RNA XIST promotes cell growth and invasion through regulating miR-497/MACC1 axis in gastric cancer. Oncotarget 8 4125–4135. 10.18632/oncotarget.13670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M., Pei Y., Wang X., Feng J., Zhang Y., Gao M. Q. (2019). LncRNA XIST mediates bovine mammary epithelial cell inflammatory response via NF-kappaB/NLRP3 inflammasome pathway. Cell Prolif. 52:e12525. 10.1111/cpr.12525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W. J., Wang H. T., Jing W., Zhou F. L., Chang L., Hong Z. F., et al. (2017). Downregulation of long non-coding RNAs JPX and XIST is associated with the prognosis of hepatocellular carcinoma. Clin. Res. Hepatol. Gas 41 163–170. 10.1016/j.clinre.2016.09.002 [DOI] [PubMed] [Google Scholar]
- Ma X., Yuan T. D., Yang C., Wang Z. S., Zang Y. J., Wu L. Q., et al. (2017). X-inactive-specific transcript of peripheral blood cells is regulated by exosomal Jpx and acts as a biomarker for female patients with hepatocellular carcinoma. Ther. Adv. Med. Oncol. 9 665–677. 10.1177/1758834017731052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Liu X., Zhu B., Li W., He Y., Cheng Z. (2020). Long noncoding RNA XIST knockdown suppresses the growth of colorectal cancer cells via regulating microRNA-338-3p/PAX5 axis. OncoTargets Ther. 596 132–142. 10.1097/cej.0000000000000596 [DOI] [PubMed] [Google Scholar]
- Maduro C., de Hoon B., Gribnau J. (2016). Fitting the puzzle pieces: the bigger picture of XCI. Trends Biochem. Sci. 41 138–147. 10.1016/j.tibs.2015.12.003 [DOI] [PubMed] [Google Scholar]
- Makhlouf M., Ouimette J. F., Oldfield A., Navarro P., Neuillet D., Rougeulle C. (2014). A prominent and conserved role for YY1 in Xist transcriptional activation. Nat. Commun. 5 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S., Inoue K., Kohda T., Sugimoto M., Mizutani E., Ogonuki N., et al. (2011). RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos. Proc. Natl. Acad. Sci. U.S.A. 108 20621–20626. 10.1073/pnas.1112664108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFaline-Figueroa J. R., Wen P. Y. (2017). The viral connection to glioblastoma. Curr. Infect. Dis. Rep. 19 1–5. [DOI] [PubMed] [Google Scholar]
- McHugh C. A., Chen C. K., Chow A., Surka C. F., Tran C., McDonel P., et al. (2015). The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521 232–236. 10.1038/nature14443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina M. A., Oza G., Sharma A., Arriaga L. G., Hernandez Hernandez J. M., Rotello V. M., et al. (2020). Triple-negative breast cancer: a review of conventional and advanced therapeutic strategies. Int. J. Environ. Res. Public Health 17 2078–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minajigi A., Froberg J. E., Wei C. Y., Sunwoo H., Kesner B., Colognori D., et al. (2015). A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 349 aab22761–aab227612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira-Bontenbal H., Gribnau J. (2016). New Xist-interacting proteins in X-chromosome inactivation. Curr. Biol. 26 338–342. [DOI] [PubMed] [Google Scholar]
- Mo Y., Lu Y., Wang P., Huang S., He L., Li D., et al. (2017). Long non-coding RNA XIST promotes cell growth by regulating miR-139-5p/PDK1/AKT axis in hepatocellular carcinoma. Tumour Biol. 39:1010428317690999. [DOI] [PubMed] [Google Scholar]
- Moga M. A., Balan A., Anastasiu C. V., Dimienescu O. G., Neculoiu C. D., Gavris C. (2018). An overview on the anticancer activity of azadirachta indica (Neem) in gynecological cancers. Int. J. Mol. Sci. 19 3989–4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monfort A., Di Minin G., Postlmayr A., Freimann R., Arieti F., Thore S., et al. (2015). Identification of spen as a crucial factor for xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep. 12 554–561. 10.1016/j.celrep.2015.06.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monfort A., Wutz A. (2020). The B-side of Xist. F1000Research 9 1–12. 10.1109/tmag.2013.2278570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montalbano A., Canver M. C., Sanjana N. E. (2017). High-throughput approaches to pinpoint function within the noncoding genome. Mol. Cell 68 44–59. 10.1016/j.molcel.2017.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller D. A., Silvan U. (2019). On the biomechanical properties of osteosarcoma cells and their environment. Int. J. Dev. Biol. 63 1–8. 10.1387/ijdb.190019us [DOI] [PubMed] [Google Scholar]
- Mutzel V., Schulz E. G. (2020). Dosage sensing, threshold responses, and epigenetic memory: a systems biology perspective on random X-chromosome inactivation, bioessays : news and reviews in molecular. Cell. Dev. Biol. 42:e1900163. [DOI] [PubMed] [Google Scholar]
- Nesterova T. B., Wei G. F., Coker H., Pintacuda G., Bowness J. S., Zhang T. Y., et al. (2019). Systematic allelic analysis defines the interplay of key pathways in X chromosome inactivation. Nat. Commun. 10 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu S., Xiang F., Jia H. (2020). Downregulation of lncRNA XIST promotes proliferation and differentiation, limits apoptosis of osteoblasts through regulating miR-203-3p/ZFPM2 axis. Nat. Methods [Epub ahead of print]. 10.1080/03008207.2020.1752200 [DOI] [PubMed] [Google Scholar]
- Ojha N., Dhamoon A. S. (2020). Myocardial Infarction. Treasure Island, FL: StatPearls. [Google Scholar]
- Pan B., Lin X., Zhang L., Hong W., Zhang Y. (2019). Long noncoding RNA X-inactive specific transcript promotes malignant melanoma progression and oxaliplatin resistance. Melanoma Res. 29 254–262. 10.1097/cmr.0000000000000560 [DOI] [PubMed] [Google Scholar]
- Patrat C., Ouimette J. F., Rougeulle C. (2020). X chromosome inactivation in human development. Development 147:dev183095. [DOI] [PubMed] [Google Scholar]
- Peng H., Luo Y. X., Ying Y. J. (2020). lncRNA XIST attenuates hypoxia-induced H9c2 cardiomyocyte injury by targeting the miR-122-5p/FOXP2 axis. Mol. Cell. Probes 50 101500–101509. 10.1016/j.mcp.2019.101500 [DOI] [PubMed] [Google Scholar]
- Pereira D., Ramos E., Branco J. (2015). Osteoarthritis. Acta Med. Port 28 99–106. [DOI] [PubMed] [Google Scholar]
- Pinheiro I., Heard E. (2017). X chromosome inactivation: new players in the initiation of gene silencing. F1000Research 6 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintacuda G., Wei G. F., Roustan C., Kirmizitas B. A., Solcan N., Cerase A., et al. (2017a). hnRNPK recruits PCGF3/5-PRC1 to the Xist RNA B-repeat to establish polycomb-mediated chromosomal silencing. Mol. Cell 68 955–969. 10.1016/j.molcel.2017.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintacuda G., Young A. N., Cerase A. (2017b). Function by structure: spotlights on Xist long non-coding RNA. Front. Mol. Biosci. 4:90. 10.3389/fmolb.2017.00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polito C., Pannuti A., Lucchesi J. C. (1990). Dosage compensation in Drosophila melanogaster male and female embryos generated by segregation distortion of the sex chromosomes. Dev. Genet. 11 249–253. 10.1002/dvg.1020110402 [DOI] [PubMed] [Google Scholar]
- Postlmayr A., Dumeau C. E., Wutz A. (2020). Cdk8 is required for establishment of H3K27me3 and gene repression by Xist and mouse development. Development 147 175141–175151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat A., Pineda E., Adamo B., Galvan P., Fernandez A., Gaba L., et al. (2015). Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast 24 26–35. [DOI] [PubMed] [Google Scholar]
- Qiu H. B., Yang K., Yu H. Y., Liu M. (2019). Downregulation of long non-coding RNA XIST inhibits cell proliferation, migration, invasion and EMT by regulating miR-212-3p/CBLL1 axis in non-small cell lung cancer cells. Eur. Rev. Med. Pharmacol. 23 8391–8402. [DOI] [PubMed] [Google Scholar]
- Quinn J. J., Chang H. Y. (2016). Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17 47–62. 10.1038/nrg.2015.10 [DOI] [PubMed] [Google Scholar]
- Richard J. L. C., Eichhorn P. J. A. (2018). Deciphering the roles of lncRNAs in breast development and disease. Oncotarget 9 20179–20212. 10.18632/oncotarget.24591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romagnolo A. P. G., Romagnolo D. F., Selmin O. I. (2015). BRCA1 as target for breast cancer prevention and therapy. Anti Cancer Agent. Meas. 15 4–14. 10.2174/1871520614666141020153543 [DOI] [PubMed] [Google Scholar]
- Ronco C., Bellomo R., Kellum J. A. (2019). Acute kidney injury. Lancet 394 1949–1964. [DOI] [PubMed] [Google Scholar]
- Rong H., Chen B., Wei X., Peng J., Ma K., Duan S., et al. (2020). Long non-coding RNA XIST expedites lung adenocarcinoma progression through upregulating MDM2 expression via binding to miR-363-3p. Thorac. Cancer 11 659–671. 10.1111/1759-7714.13310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropers H. H., Wolff G., Hitzeroth H. W. (1978). Preferential X inactivation in human placenta membranes: is the paternal X inactive in early embryonic development of female mammals? Hum. Genet. 43 265–273. 10.1007/bf00278833 [DOI] [PubMed] [Google Scholar]
- Sado T., Brockdorff N. (2013). Advances in understanding chromosome silencing by the long non-coding RNA Xist. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 368:20110325. 10.1098/rstb.2011.0325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahakyan A., Yang Y., Plath K. (2018). The role of xist in X-chromosome dosage compensation. Trends Cell Biol. 28 999–1013. 10.1016/j.tcb.2018.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi T., Hasegawa Y., Brockdorff N., Tsutsui K., Tsutsui K. M., Sado T., et al. (2016). Control of chromosomal localization of Xist by hnRNP U family molecules. Dev. Cell. 39 11–12. 10.1016/j.devcel.2016.09.022 [DOI] [PubMed] [Google Scholar]
- Salama E. A., Adbeltawab R. E., El Tayebi H. M. (2019). XIST and TSIX: novel cancer immune biomarkers in PD-L1-overexpressing breast cancer patients. Front. Oncol. 9:1459. 10.3389/fonc.2019.01459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador M. A., Wicinski J., Cabaud O., Toiron Y., Finetti P., Josselin E., et al. (2013). The histone deacetylase inhibitor abexinostat induces cancer stem cells differentiation in breast cancer with low Xist expression. Clin. Cancer Res. 19 6520–6531. 10.1158/1078-0432.ccr-13-0877 [DOI] [PubMed] [Google Scholar]
- Sandoval Y., Smith S. W., Sexter A., Thordsen S. E., Bruen C. A., Carlson M. D., et al. (2017). Type 1 and 2 myocardial infarction and myocardial injury: clinical transition to high-sensitivity cardiac troponin I. Am. J. Med. 130 1431–1439. [DOI] [PubMed] [Google Scholar]
- Sandoval Y., Thygesen K. (2017). Myocardial infarction type 2 and myocardial injury. Clin. Chem. 63 101–107. 10.1373/clinchem.2016.255521 [DOI] [PubMed] [Google Scholar]
- Sarma K., Cifuentes-Rojas C., Ergun A., del Rosario A., Jeon Y., White F., et al. (2014). ATRX directs binding of PRC2 to Xist RNA and polycomb targets. Cell 159 869–883. 10.1016/j.cell.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadendorf D., van Akkooi A. C. J., Berking C., Griewank K. G., Gutzmer R., Hauschild A., et al. (2018). Melanoma. Lancet 392 971–984. [DOI] [PubMed] [Google Scholar]
- Scheltens P., Blennow K., Breteler M. M. B., de Strooper B., Frisoni G. B., Salloway S., et al. (2016). Alzheimer’s disease. Lancet 388 505–517. [DOI] [PubMed] [Google Scholar]
- Schertzer M. D., Braceros K. C. A., Starmer J., Cherney R. E., Lee D. M., Salazar G., et al. (2019). lncRNA-induced spread of polycomb controlled by genome architecture, RNA abundance, and CpG Island DNA. Mol. Cell 75 523–537. 10.1016/j.molcel.2019.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz S. U., Grote P., Herrmann B. G. (2016). Mechanisms of long noncoding RNA function in development and disease. Cell Mol. Life Sci. 73 2491–2509. 10.1007/s00018-016-2174-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten P. C., Vollebergh M. A., Opdam M., Jonkers M., Loden M., Wesseling J., et al. (2016). High XIST and low 53BP1 expression predict poor outcome after high-dose alkylating chemotherapy in patients with a BRCA1-like breast cancer. Mol. Cancer Ther. 15 190–198. 10.1158/1535-7163.mct-15-0470 [DOI] [PubMed] [Google Scholar]
- Shen C., Li J. (2020). LncRNA XIST silencing protects against sepsis-induced acute liver injury via inhibition of BRD4 expression. Inflammation 1321 1–12. [DOI] [PubMed] [Google Scholar]
- Shen J., Hong L., Yu D., Cao T., Zhou Z., He S. (2019). LncRNA XIST promotes pancreatic cancer migration, invasion and EMT by sponging miR-429 to modulate ZEB1 expression. Int. J. Biochem. Cell Biol. 113 17–26. 10.1016/j.biocel.2019.05.021 [DOI] [PubMed] [Google Scholar]
- Shen J., Xiong J., Shao X., Cheng H., Fang X., Sun Y., et al. (2020). Knockdown of the long noncoding RNA XIST suppresses glioma progression by upregulating miR-204-5p. J. Cancer 11 4550–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X. F., Cheng Y., Dong Q. R., Zheng M. Q. (2020). MicroRNA-675-3p regulates IL-1β-stimulated human chondrocyte apoptosis and cartilage degradation by targeting GNG5. Biochem. Biophys. Res. Commun. 527 458–465. 10.1016/j.bbrc.2020.04.044 [DOI] [PubMed] [Google Scholar]
- Shenoda B. B., Ramanathan S., Gupta R., Tian Y., Jean-Toussaint R., Alexander G. M., et al. (2020). Xist attenuates acute inflammatory response by female cells. Cell Mol. Life Sci. 2020 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoda B. B., Tian Y. Z., Alexander G. M., Aradillas-Lopez E., Schwartzman R. J., Ajit S. K. (2018). miR-34a-mediated regulation of XIST in female cells under inflammation. J. Pain Res. 11 935–945. 10.2147/jpr.s159458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherstyuk V. V., Shevchenko A. I., Mazurok N. A., Zakian S. M. (2013). Replication origin activity in vole X chromosome inactivation center in different cell types, Doklady. Biochem. Biophys. 450 164–166. 10.1134/s1607672913030125 [DOI] [PubMed] [Google Scholar]
- Shi J. F., Tan S. L., Song L. M., Song L. S., Wang Y. S. (2020). LncRNA XIST knockdown suppresses the malignancy of human nasopharyngeal carcinoma through XIST/miRNA-148a-3p/ADAM17 pathway in vitro and in vivo. Biomed. Pharmacother. 121:109620. 10.1016/j.biopha.2019.109620 [DOI] [PubMed] [Google Scholar]
- Shi X., Sun M., Liu H., Yao Y., Song Y. (2013). Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 339 159–166. 10.1016/j.canlet.2013.06.013 [DOI] [PubMed] [Google Scholar]
- Shukla S., Penta D., Mondal P., Meeran S. M. (2019). Epigenetics of breast cancer: clinical status of epi-drugs and phytochemicals. Adv. Exp. Med. Biol. 1152 293–310. 10.1007/978-3-030-20301-6_16 [DOI] [PubMed] [Google Scholar]
- Sidorenko J., Kassam I., Kemper K. E., Zeng J., Lloyd-Jones L. R., Montgomery G. W., et al. (2019). The effect of X-linked dosage compensation on complex trait variation. Nat. Commun. 10 3009–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R. L., Miller K. D., Jemal A. (2019). Cancer statistics, 2019. CA Cancer J. Clin. 69 7–34. [DOI] [PubMed] [Google Scholar]
- Siegel R. L., Miller K. D., Jemal A. (2020). Cancer statistics, 2020. CA Cancer J. Clin. 70 7–30. [DOI] [PubMed] [Google Scholar]
- Sirchia S. M., Ramoscelli L., Grati F. R., Barbera F., Coradini D., Rossella F., et al. (2005). Loss of the inactive X chromosome and replication of the active x in BRCA1-defective and wild-type breast cancer cells. Cancer Res. 65 2139–2146. 10.1158/0008-5472.can-04-3465 [DOI] [PubMed] [Google Scholar]
- Sirchia S. M., Tabano S., Monti L., Recalcati M. P., Gariboldi M., Grati F. R., et al. (2009). Misbehaviour of XIST RNA in breast cancer cells. PLoS One 4:e5559. 10.1371/journal.pone.0005559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack F. J., Chinnaiyan A. M. (2019). The role of non-coding RNAs in oncology. Cell 179 1033–1055. 10.1016/j.cell.2019.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrabifar N. (2020). Variation in the expression level of MALAT1, MIAT and XIST lncRNAs in coronary artery disease patients with and without type 2 diabetes mellitus. Cell Biol. Int. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Song H., He P., Shao T. S., Li Y., Li J., Zhang Y. J. (2017). Long non-coding RNA XIST functions as an oncogene in human colorectal cancer by targeting miR-132-3p. J. Buon 22 696–703. [PubMed] [Google Scholar]
- Song P., Ye L. F., Zhang C., Peng T., Zhou X. H. (2016). Long non-coding RNA XIST exerts oncogenic functions in human nasopharyngeal carcinoma by targeting miR-34a-5p. Gene 592 8–14. 10.1016/j.gene.2016.07.055 [DOI] [PubMed] [Google Scholar]
- Soudyab M., Iranpour M., Ghafouri-Fard S. (2016). The role of long non-coding RNAs in breast cancer. Arch. Iranian Med. 19 508–517. [PubMed] [Google Scholar]
- Spatz A., Borg C., Feunteun J. (2004). X-chromosome genetics and human cancer. Nat. Rev. Cancer 4 617–629. [DOI] [PubMed] [Google Scholar]
- Sripathy S., Leko V., Adrianse R. L., Loe T., Foss E. J., Dalrymple E., et al. (2017). Screen for reactivation of MeCP2 on the inactive X chromosome identifies the BMP/TGF-beta superfamily as a regulator of XIST expression. Proc. Natl. Acad. Sci. U.S.A. 114 1619–1624. 10.1073/pnas.1621356114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Laurent G., Wahlestedt C., Kapranov P. (2015). The Landscape of long noncoding RNA classification. Trends Genet. 31 239–251. 10.1016/j.tig.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuer C. E., El-Deiry M., Parks J. R., Higgins K. A., Saba N. F. (2017). An update on larynx cancer. CA Cancer J. Clin. 67 31–50. 10.3322/caac.21386 [DOI] [PubMed] [Google Scholar]
- Stewart C., Ralyea C., Lockwood S. (2019). Ovarian cancer: an integrated review. Semin. Oncol. Nurs. 35 151–156. [DOI] [PubMed] [Google Scholar]
- Strehle M., Guttman M. (2020). Xist drives spatial compartmentalization of DNA and protein to orchestrate initiation and maintenance of X inactivation. Mol. Genet. Genom. Med. 64 139–147. 10.1016/j.ceb.2020.04.009 [DOI] [PubMed] [Google Scholar]
- Sun J., Pan L. M., Chen L. B., Wang Y. (2017). LncRNA XIST promotes human lung adenocarcinoma cells to cisplatin resistance via let-7i/BAG-1 axis. Cell Cycle 16 2100–2107. 10.1080/15384101.2017.1361071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J. M., Zhang Y. B. (2019). LncRNA XIST enhanced TGF-beta 2 expression by targeting miR-141-3p to promote pancreatic cancer cells invasion. Bioscience Rep. 39:BSR20190332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K., Jia Z., Duan R., Yan Z., Jin Z., Yan L., et al. (2019). Long non-coding RNA XIST regulates miR-106b-5p/P21 axis to suppress tumor progression in renal cell carcinoma. Biochem. Biophys. Res. Commun. 510 416–420. 10.1016/j.bbrc.2019.01.116 [DOI] [PubMed] [Google Scholar]
- Sun N. N., Zhang G. Z., Liu Y. Y. (2018). Long non-coding RNA XIST sponges miR-34a to promotes colon cancer progression via Wnt/beta-catenin signaling pathway. Gene 665 141–148. 10.1016/j.gene.2018.04.014 [DOI] [PubMed] [Google Scholar]
- Sun W., Zu Y. K., Fu X. N., Deng Y. (2017). Knockdown of lncRNA-XIST enhances the chemosensitivity of NSCLC cells via suppression of autophagy. Oncol. Rep. 38 3347–3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W. B., Ma M. N., Yu H. M., Yu H. (2018). Inhibition of lncRNA X inactivate-specific transcript ameliorates inflammatory pain by suppressing satellite glial cell activation and inflammation by acting as a sponge of miR-146a to inhibit Na(v)1.7. J. Cell. Biochem. 119 9888–9898. 10.1002/jcb.27310 [DOI] [PubMed] [Google Scholar]
- Sun X., Wei B., Peng Z. H., Fu Q. L., Wang C. J., Zheng J. C., et al. (2019). Knockdown of lncRNA XIST suppresses osteosarcoma progression by inactivating AKT/mTOR signaling pathway by sponging miR-375-3p. Int. J. Clin. Exp. Pathol. 12 1507–1517. [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Lv B., Zhang X. (2020). Knock-down of LncRNA-XIST induced glioma cell death and inhibited tumorigenesis by regulating miR-137/SLC1A5 axis-mediated ROS production. Naunyn Schmiedebergs Arch. Pharmacol. 394:557. 10.1007/s00210-020-01831-3 [DOI] [PubMed] [Google Scholar]
- Sun Y. B., Xu J. J. (2019). TCF-4 regulated lncRNA-XIST promotes M2 polarization of macrophages and is associated with lung cancer. OncoTargets Ther. 12 8055–8062. 10.2147/ott.s210952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Zhang B., Cui T. (2018). Long non-coding RNA XIST exerts oncogenic functions in pancreatic cancer via miR-34a-5p. Oncol. Rep. 39 1591–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrett C. M., Paneru B., Sandoval-Heglund D., Wang J. L., Banerjee S., Sindhava V., et al. (2019). Altered X-chromosome inactivation in T cells may promote sex-biased autoimmune diseases. JCI Insight 4:e126751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrett C. M., Sierra I., Beethem Z. T., Dubin A. H., Anguera M. C. (2020). Loss of epigenetic modifications on the inactive X chromosome and sex-biased gene expression profiles in B cells from NZB/W F1 mice with lupus-like disease. J. Autoimmun. 107:102357. 10.1016/j.jaut.2019.102357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrett C. M., Sindhava V., Hodawadekar S., Myles A., Liang G. X., Zhang Y., et al. (2017). Loss of Xist RNA from the inactive X during B cell development is restored in a dynamic YY1-dependent two-step process in activated B cells. PLoS Genet. 13:e1007050. 10.1371/journal.pgen.1007050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B., Li W., Ji T., Li X., Qu X., Feng L., et al. (2020). Downregulation of XIST ameliorates acute kidney injury by sponging miR-142-5p and targeting PDCD4. J. Cell. Physiol. 2020 1–12. 10.1155/2021/6624702 [DOI] [PubMed] [Google Scholar]
- Tang Y., He R., An J., Deng P., Huang L., Yang W. (2017). lncRNA XIST interacts with miR-140 to modulate lung cancer growth by targeting iASPP. Oncol. Rep. 38 941–948. 10.3892/or.2017.5751 [DOI] [PubMed] [Google Scholar]
- Tantai J. C., Hu D. Z., Yang Y., Geng J. F. (2015). Combined identification of long non-coding RNA XIST and HIF1A-AS1 in serum as an effective screening for non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 8 7887–7895. [PMC free article] [PubMed] [Google Scholar]
- Thompson L. D. (2017). Laryngeal dysplasia, squamous cell carcinoma, and variants. Surg. Pathol. Clin. 10 15–33. 10.1016/j.path.2016.10.003 [DOI] [PubMed] [Google Scholar]
- Thorvaldsen J. L., Weaver J. R., Bartolomei M. S. (2011). A YY1 bridge for X inactivation. Cell 146 11–13. 10.1016/j.cell.2011.06.029 [DOI] [PubMed] [Google Scholar]
- Tian K., Sun D., Chen M., Yang Y., Wang F., Guo T., et al. (2020). Long Noncoding RNA X-inactive specific transcript facilitates cellular functions in melanoma via miR-139-5p/ROCK1 pathway. OncoTargets Ther. 13 1277–1287. 10.2147/ott.s225661 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tian L. J., Wu Y. P., Wang D., Zhou Z. H., Shou B., Zhang D. Y., et al. (2019). Upregulation of long noncoding RNA (lncRNA) X-inactive specific transcript (XIST) is associated with cisplatin resistance in non-small cell lung cancer (NSCLC) by downregulating MicroRNA-144-3p. Med. Sci. Monit. 25 8095–8104. 10.12659/msm.916075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian R., Wang L. F., Zou H., Song M. J., Liu L., Zhang H. (2020). Role of the XIST-miR-181a-COL4A1 axis in the development and progression of keratoconus. Mol. Vis. 26 1–13. [PMC free article] [PubMed] [Google Scholar]
- van Bemmel J. G., Galupa R., Gard C., Servant N., Picard C., Davies J., et al. (2019). The bipartite TAD organization of the X-inactivation center ensures opposing developmental regulation of Tsix and Xist. Nat. Genet. 51 1024–1034. 10.1038/s41588-019-0412-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bemmel J. G., Mira-Bontenbal H., Gribnau J. (2016). Cis- and trans-regulation in X inactivation. Chromosoma 125 41–50. 10.1007/s00412-015-0525-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velázquez-Flores M., Rodríguez-Corona J. M., ópez-Aguilar J. E. L., Siordia-Reyes G., Ramírez-Reyes G., ánchez-Rodríguez G. S., et al. (2020). Noncoding RNAs as potential biomarkers for DIPG diagnosis and prognosis: XIST and XIST-210 involvement. Clin. Transl. Oncol. 7 1–13. 10.1155/2020/8822859 [DOI] [PubMed] [Google Scholar]
- Vincent-Salomon A., Ganem-Elbaz C., Manie E., Raynal V., Sastre-Garau X., Stoppa-Lyonnet D., et al. (2007). X inactive-specific transcript RNA coating and genetic instability of the X chromosome in BRCA1 breast tumors. Cancer Res. 67 5134–5140. 10.1158/0008-5472.can-07-0465 [DOI] [PubMed] [Google Scholar]
- Waldron D. (2016). Xist as a recruitment tool. Nat. Rev. Genet. 17:1. [DOI] [PubMed] [Google Scholar]
- Wang C. H., Qi S., Xie C., Li C. F., Wang P., Liu D. M. (2018a). Upregulation of long non-coding RNA XIST has anticancer effects on epithelial ovarian cancer cells through inverse downregulation of hsa-miR-214-3p. J. Gynecol. Oncol. 29:e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. Y., Colognori D., Sunwoo H., Wang D. N., Lee J. T. (2019a). PRC1 collaborates with SMCHD1 to fold the X-chromosome and spread Xist RNA between chromosome compartments. Nat. Commun. 10 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Tang L., Wu Y., Fan C., Zhang S., Xiang B., et al. (2020a). Abnormal X chromosome inactivation and tumor development. Cell Mol. Life Sci. 77 2949–2958. 10.1007/s00018-020-03469-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Li W., Tan G. (2019b). Long non-coding RNA XIST modulates cisplatin resistance by altering PDCD4 and Fas-Lexpressions in human nasopharyngeal carcinoma HNE1 cells in vitro. Nan Fang Yi Ke Da Xue Xue Bao 39 357–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. Y., Shen Q. M., Zhang X., Yang C. L., Cui S., Sun Y. B., et al. (2017). The long non-coding RNA XIST controls non-small cell lung cancer proliferation and invasion by modulating miR-186-5p. Cell. Physiol. Biochem. 41 2221–2229. 10.1159/000475637 [DOI] [PubMed] [Google Scholar]
- Wang J. L., Cai H. B., Dai Z. X., Wang G. (2019c). Down-regulation of lncRNA XIST inhibits cell proliferation via regulating miR-744/RING1 axis in non-small cell lung cancer. Clin. Sci. 133 1567–1579. 10.1042/cs20190519 [DOI] [PubMed] [Google Scholar]
- Wang Q. (2020). XIST silencing alleviated inflammation and mesangial cells proliferation in diabetic nephropathy by sponging miR-485. Arch. Physiol. Biochem. 2020 1–7. 10.1080/13813455.2020.1789880 [DOI] [PubMed] [Google Scholar]
- Wang Q. Y., Peng L., Chen Y., Liao L. D., Chen J. X., Li M., et al. (2020b). Characterization of super-enhancer-associated functional lncRNAs acting as ceRNAs in ESCC. Development 14 2203–2230. 10.1002/1878-0261.12726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. G., Cao F., Gu X. S., Chen J. N., Xu R. X., Huang Y. N., et al. (2019d). LncRNA XIST, as a ceRNA of miR-204, aggravates lipopolysaccharide-induced acute respiratory distress syndrome in mice by upregulating IRF2. Int. J. Clin. Exp. Pathol. 12 2425–2434. [PMC free article] [PubMed] [Google Scholar]
- Wang S. L., Li G. Z. (2020). LncRNA XIST inhibits ovarian cancer cell growth and metastasis via regulating miR-150-5p/PDCD4 signaling pathway. N. S. Arch. Pharmacol. 13 1–13. [DOI] [PubMed] [Google Scholar]
- Wang T., Liu Y., Wang Y., Huang X., Zhao W., Zhao Z. (2019e). Long non-coding RNA XIST promotes extracellular matrix degradation by functioning as a competing endogenous RNA of miR-1277-5p in osteoarthritis. Int. J. Mol. Med. 44 630–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Shen H., Cao G., Huang J. (2019f). Long non-coding RNA XIST predicts poor prognosis and promotes malignant phenotypes in osteosarcoma. Oncol. Lett. 17 256–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang C., Geng C., Zhao K. (2018b). LncRNA XIST knockdown attenuates Abeta25-35-induced toxicity, oxidative stress, and apoptosis in primary cultured rat hippocampal neurons by targeting miR-132. Int. J. Clin. Exp. Pathol. 11 3915–3924. [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhang G. J., Cheng Z., Dai L. L., Jia L. Q., Jing X. G., et al. (2018c). Knockdown of LncRNA-XIST suppresses proliferation and TGF-beta 1-induced EMT in NSCLC through the Notch-1 pathway by regulation of miR-137. Genet. Test. Mol. Biomark. 22 333–342. 10.1089/gtmb.2018.0026 [DOI] [PubMed] [Google Scholar]
- Wang Y., Sun D., Sheng Y., Guo H., Meng F., Song T. (2020c). XIST promotes cell proliferation and invasion by regulating miR-140-5p and SOX4 in retinoblastoma. World J. Surg. Oncol. 18 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. C., Liang Y., Luo J. M., Nie J., Yin H. M., Chen Q., et al. (2017). XIST/miR-139 axis regulates bleomycin (BLM)-induced extracellular matrix (ECM) and pulmonary fibrosis through beta-catenin. Oncotarget 8 65359–65369. 10.18632/oncotarget.18310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Yuan J., Li L., Yang Y., Xu X., Wang Y. (2017). Long non-coding RNA XIST exerts oncogenic functions in human glioma by targeting miR-137. Am. J. Transl. Res. 9 1845–1855. [PMC free article] [PubMed] [Google Scholar]
- Wang Z. Q., Xiu D. H., Jiang J. L., Liu G. F. (2020d). Long non-coding RNA XIST binding to let-7c-5p contributes to rheumatoid arthritis through its effects on proliferation and differentiation of osteoblasts via regulation of STAT3. Nature 2020:e23496. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang Z. Y., Qiu H., He J. B., Liu L. X., Xue W., Fox A., et al. (2019g). The emerging roles of hnRNPK. J. Cell. Physiol. 235 1–14. [DOI] [PubMed] [Google Scholar]
- Waśko U., Zheng Z., Bhatnagar S. (2019). Visualization of Xist long noncoding RNA with a fluorescent CRISPR/Cas9 system. Epitranscriptomics 2019 41–50. 10.1007/978-1-4939-8808-2_3 [DOI] [PubMed] [Google Scholar]
- Webb P. M., Jordan S. J. (2017). Epidemiology of epithelial ovarian cancer, Best practice & research. Clin. Obstetr. Gynaecol. 41 3–14. 10.1016/j.bpobgyn.2016.08.006 [DOI] [PubMed] [Google Scholar]
- Wehbe N., Nasser S. A., Pintus G., Badran A., Eid A. H., Baydoun E. (2019). MicroRNAs in cardiac hypertrophy. Int. J. Mol. Sci. 20 4714–4731. 10.3390/ijms20194714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K. R., Zheng R. S., Zhang S. W., Liang Z. H., Li Z. M., Chen W. Q. (2017). Nasopharyngeal carcinoma incidence and mortality in China, 2013. Chinese J. Cancer 36 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M., Li L., Zhang Y., Zhang Z. J., Liu H. L., Bao H. G. (2019). LncRNA X inactive specific transcript contributes to neuropathic pain development by sponging miR-154-5p via inducing toll-like receptor 5 in CCI rat models. J. Cell. Biochem. 120 1271–1281. 10.1002/jcb.27088 [DOI] [PubMed] [Google Scholar]
- Wei W., Liu Y., Lu Y. B., Yang B., Tang L. (2017). LncRNA XIST promotes pancreatic cancer proliferation through miR-133a/EGFR. J. Cell. Biochem. 118 3349–3358. 10.1002/jcb.25988 [DOI] [PubMed] [Google Scholar]
- Wen J. F., Jiang Y. Q., Li C., Dai X. K., Wu T., Yin W. Z. (2020). LncRNA-XIST promotes the oxidative stress-induced migration, invasion, and epithelial-to-mesenchymal transition of osteosarcoma cancer cells through miR-153-SNAI1 axis. Cell Biol. Int. 10 1991–2001. 10.3389/fonc.2020.577280 [DOI] [PubMed] [Google Scholar]
- Willard H. F. (1996). X chromosome inactivation, XIST, and pursuit of the X-inactivation center. Cell 86 5–7. 10.1016/s0092-8674(00)80071-9 [DOI] [PubMed] [Google Scholar]
- Wu D. P., Nie X. G., Ma C., Liu X. H., Liang X., An Y. B., et al. (2017). RSF1 functions as an oncogene in osteosarcoma and is regulated by XIST/miR-193a-3p axis. Biomed. Pharmacother. 95 207–214. 10.1016/j.biopha.2017.08.068 [DOI] [PubMed] [Google Scholar]
- Wu X., Dinglin X., Wang X., Luo W., Shen Q., Li Y., et al. (2017). Long noncoding RNA XIST promotes malignancies of esophageal squamous cell carcinoma via regulation of miR-101/EZH2. Oncotarget 8 76015–76028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao D., Cui X., Wang X. (2019). Long noncoding RNA XIST increases the aggressiveness of laryngeal squamous cell carcinoma by regulating miR-124-3p/EZH2. Exp. Cell Res. 381 172–178. 10.1016/j.yexcr.2019.04.034 [DOI] [PubMed] [Google Scholar]
- Xiao L., Gu Y., Sun Y., Chen J., Wang X., Zhang Y., et al. (2019). The long noncoding RNA XIST regulates cardiac hypertrophy by targeting miR-101. J. Cell. Physiol. 234 13680–13692. 10.1002/jcp.28047 [DOI] [PubMed] [Google Scholar]
- Xiao Y., Yurievich U. A., Yosypovych S. V. (2017). Long noncoding RNA XIST is a prognostic factor in colorectal cancer and inhibits 5-fluorouracil-induced cell cytotoxicity through promoting thymidylate synthase expression. Oncotarget 8 83171–83182. 10.18632/oncotarget.20487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z. Y., Wang F. F., Xiao Z. H., Liu S. F., Lai Y. L., Tang S. L. (2019). Long noncoding RNA XIST enhances ethanol-induced hepatic stellate cells autophagy and activation via miR-29b/HMGB1 axis. IUBMB Life 71 1962–1972. 10.1002/iub.2140 [DOI] [PubMed] [Google Scholar]
- Xing F., Liu Y., Wu S. Y., Wu K., Sharma S., Mo Y. Y., et al. (2018). Loss of XIST in breast cancer activates MSN-c-met and reprograms microglia via exosomal miRNA to promote brain metastasis. Cancer Res. 78 4316–4330. 10.1158/0008-5472.can-18-1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Wang L., Li Y., Chen M., He W., Qi L. (2017). The long non-coding RNA XIST interacted with MiR-124 to modulate bladder cancer growth, invasion and migration by targeting androgen receptor (AR). Cell. Physiol. Biochem. 43 405–418. 10.1159/000480419 [DOI] [PubMed] [Google Scholar]
- Xu G. H., Mo L. J., Wu C. N., Shen X. Y., Dong H. L., Yu L. F., et al. (2019). The miR-15a-5p-XIST-CUL3 regulatory axis is important for sepsis-induced acute kidney injury. Renal Fail. 41 955–966. 10.1080/0886022x.2019.1669460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Zhang Y. G., Huang Y. G. (2016). Advances in the treatment of neuropathic pain. Transl. Res. Pain Itch 904 117–129. [DOI] [PubMed] [Google Scholar]
- Xu N., Tsai C. L., Lee J. T. (2006). Transient homologous chromosome pairing marks the onset of X inactivation. Science 311 1149–1152. 10.1126/science.1122984 [DOI] [PubMed] [Google Scholar]
- Xu R., Zhu X., Chen F., Huang C., Ai K., Wu H., et al. (2018). LncRNA XIST/miR-200c regulates the stemness properties and tumourigenicity of human bladder cancer stem cell-like cells. Cancer Cell Int. 18 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Jiang W., Fan L., Gao Q., Li G. (2017). Upregulation of long noncoding RNA Xist promotes proliferation of osteosarcoma by epigenetic silencing of P21. Oncotarget 8 101406–101417. 10.18632/oncotarget.20738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X. H., Ma C. M., Liu C., Duan Z. H., Zhang L. (2018). Knockdown of long noncoding RNA XIST alleviates oxidative low-density lipoprotein-mediated endothelial cells injury through modulation of miR-320/NOD2 axis. Biochem. Biophys. Res. Commun. 503 586–592. 10.1016/j.bbrc.2018.06.042 [DOI] [PubMed] [Google Scholar]
- Xu X. H., Zhou X. Y., Chen Z. J., Gao C., Zhao L., Cui Y. S. (2020). Silencing of lncRNA XIST inhibits non-small cell lung cancer growth and promotes chemosensitivity to cisplatin. Aging Us 12 4711–4726. 10.18632/aging.102673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Wang J., Wang J. (2018). Long noncoding RNA XIST promotes proliferation and invasion by targeting miR-141 in papillary thyroid carcinoma. OncoTargets Ther. 11 5035–5043. 10.2147/ott.s170439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z. Z., Xu J., Lu H. Q., Lin B., Cai S. P., Guo J., et al. (2017). LARP1 is regulated by the XIST/miR-374a axis and functions as an oncogene in non-small cell lung carcinoma. Oncol. Rep. 38 3659–3667. [DOI] [PubMed] [Google Scholar]
- Yan F. R., Wang X. D., Zeng Y. M. (2019). 3D genomic regulation of lncRNA and Xist in X chromosome. Semin. Cell Dev. Biol. 90 174–180. 10.1016/j.semcdb.2018.07.013 [DOI] [PubMed] [Google Scholar]
- Yan X. T., Lu J. M., Wang Y., Cheng X. L., He X. H., Zheng W. Z., et al. (2018). XIST accelerates neuropathic pain progression through regulation of miR-150 and ZEB1 in CCI rat models. J. Cell. Physiol. 233 6098–6106. 10.1002/jcp.26453 [DOI] [PubMed] [Google Scholar]
- Yang C., Wu K., Wang S., Wei G. (2018). Long non-coding RNA XIST promotes osteosarcoma progression by targeting YAP via miR-195-5p. J. Cell. Biochem. 119 5646–5656. 10.1002/jcb.26743 [DOI] [PubMed] [Google Scholar]
- Yang H., Zhang X., Zhao Y., Sun G., Zhang J., Gao Y., et al. (2020). Downregulation of lncRNA XIST represses tumor growth and boosts radiosensitivity of neuroblastoma via modulation of the miR-375/L1CAM axis. Neurochem. Res. 9 1–12. [DOI] [PubMed] [Google Scholar]
- Yang J., Shen Y., Yang X., Long Y., Chen S., Lin X., et al. (2019). Silencing of long noncoding RNA XIST protects against renal interstitial fibrosis in diabetic nephropathy via microRNA-93-5p-mediated inhibition of CDKN1A. Am. J. Physiol. Renal Physiol. 317 F1350–F1358. [DOI] [PubMed] [Google Scholar]
- Yang L. G., Cao M. Z., Zhang J., Li X. Y., Sun Q. L. (2020). LncRNA XIST modulates HIF-1A/AXL signaling pathway by inhibiting miR-93-5p in colorectal cancer. Mol. Genet. Genom. Med. 8:e1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Wei W. B. (2019). Long non-coding RNAs in retinoblastoma. Pathol. Res. Pract. 215 1–6. [DOI] [PubMed] [Google Scholar]
- Yang X., Zhang S., He C., Xue P., Zhang L., He Z., et al. (2020). METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol. Cancer 19 46–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Jiang X. D., Jiang X. F., Zhao H. Y. (2018). X-inactive-specific transcript: a long noncoding RNA with complex roles in human cancers. Gene 679 28–35. 10.1016/j.gene.2018.08.071 [DOI] [PubMed] [Google Scholar]
- Yao L., Yang L., Song H., Liu T. G., Yan H. (2020). Silencing of lncRNA XIST suppresses proliferation and autophagy and enhances vincristine sensitivity in retinoblastoma cells by sponging miR-204-5p. Eur. Rev. Med. Pharmacol. Sci. 24 3526–3537. [DOI] [PubMed] [Google Scholar]
- Yao Y., Ma J., Xue Y., Wang P., Li Z., Liu J., et al. (2015). Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 359 75–86. 10.1016/j.canlet.2014.12.051 [DOI] [PubMed] [Google Scholar]
- Yi W., Li J. (2020). CRISPR-assisted detection of RNA-protein interactions in living cells. eLife 17 685–688. 10.1038/s41592-020-0866-0 [DOI] [PubMed] [Google Scholar]
- Yin Y., Lu J. Y., Zhang X., Shao W., Xu Y., Li P., et al. (2020). U1 snRNP regulates chromatin retention of noncoding RNAs. Nature 580 147–150. 10.1038/s41586-020-2105-3 [DOI] [PubMed] [Google Scholar]
- Yu B., van Tol H. T. A., Stout T. A. E. (2020). Initiation of X chromosome inactivation during bovine embryo development. Cells Basel 9 1016–1034. 10.3390/cells9041016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Xue Y., Wang P., Liu X., Ma J., Zheng J., et al. (2017). Knockdown of long non-coding RNA XIST increases blood-tumor barrier permeability and inhibits glioma angiogenesis by targeting miR-137. Oncogenesis 6:e303. 10.1038/oncsis.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Dong W., Liang J. (2020). Extracellular vesicle-transported long non-coding RNA (LncRNA) X inactive-specific transcript (XIST) in serum is a potential novel biomarker for colorectal cancer diagnosis. Med. Sci. Monit. 26:e924448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N. H., Liang Y., Zhu H., Mo H. Y., Pei H. P. (2017). CsA promotes XIST expression to regulate human trophoblast cells proliferation and invasion through miR-144/Titin axis. J. Cell. Biochem. 118 2208–2218. 10.1002/jcb.25867 [DOI] [PubMed] [Google Scholar]
- Yue M. H., Ogawa Y. (2018). CRISPR/Cas9-mediated modulation of splicing efficiency reveals short splicing isoform of Xist RNA is sufficient to induce X-chromosome inactivation. Nucleic Acids Res. 46:e26. 10.1093/nar/gkx1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Cao Z., Ding X., Wei X., Zhang X., Hou J., et al. (2017). The lncRNA XIST regulates the tumorigenicity of renal cell carcinoma cells via the miR-302c/SDC1 axis. Int. J. Clin. Exp. Pathol. 10 7481–7491. [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Li W. Y., Yang Y., Yan L. Z., Zhang S. Y., He J., et al. (2019a). LncRNA XIST facilitates cell growth, migration and invasion via modulating H3 histone methylation of DKK1 in neuroblastoma. Cell Cycle 18 1882–1892. 10.1080/15384101.2019.1632134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. D., Gao X. F., Yang J., Fan X. Y., Wang W., Liang Y. F., et al. (2019b). Xist intron 1 repression by transcriptional-activator-like effectors designer transcriptional factor improves somatic cell reprogramming in mice. Stem Cells 37 599–608. 10.1002/stem.2928 [DOI] [PubMed] [Google Scholar]
- Zhang M., Liu H. Y., Han Y. L., Wang L., Zhai D. D., Ma T., et al. (2019c). Silence of lncRNA XIST represses myocardial cell apoptosis in rats with acute myocardial infarction through regulating miR-449. Eur. Rev. Med. Pharmacol. Sci. 23 8566–8572. [DOI] [PubMed] [Google Scholar]
- Zhang M., Wang F., Xiang Z., Huang T., Zhou W. B. (2020a). LncRNA XIST promotes chemoresistance of breast cancer cells to doxorubicin by sponging miR-200c-3p to upregulate ANLN. Clin. Exp. Pharmacol. Physiol. 47 1464–1472. 10.1111/1440-1681.13307 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Chen B. Y., Liu P., Yang J. (2018). XIST promotes gastric cancer (GC) progression through TGF-beta 1 via targeting miR-185. J. Cell. Biochem. 119 2787–2796. 10.1002/jcb.26447 [DOI] [PubMed] [Google Scholar]
- Zhang R., Wang Z., Yu Q., Shen J., He W., Zhou D., et al. (2019d). Atractylenolide II reverses the influence of lncRNA XIST/miR-30a-5p/ROR1 axis on chemo-resistance of colorectal cancer cells. J. Cell. Mol. Med. 23 3151–3165. 10.1111/jcmm.14148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Xia T. (2017). Long non-coding RNA XIST regulates PDCD4 expression by interacting with miR-21-5p and inhibits osteosarcoma cell growth and metastasis. Int. J. Oncol. 51 1460–1470. 10.3892/ijo.2017.4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wu H., Mai C., Qi Y. (2020b). Long noncoding RNA XIST/miR-17/PTEN axis modulates the proliferation and apoptosis of vascular smooth muscle cells to affect stanford type A aortic dissection. J. Cardiovasc. Pharmacol. 76 53–62. 10.1097/fjc.0000000000000835 [DOI] [PubMed] [Google Scholar]
- Zhang X. T., Pan S. X., Wang A. H., Kong Q. Y., Jiang K. T., Yu Z. B. (2019e). Long non-coding RNA (lncRNA) X-inactive specific transcript (XIST) plays a critical role in predicting clinical prognosis and progression of colorectal cancer. Med. Sci. Monit. 25 6429–6435. 10.12659/msm.915329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li X., Gibson A., Edberg J., Kimberly R. P., Absher D. M. (2020c). Skewed allelic expression on X chromosome associated with aberrant expression of XIST on systemic lupus erythematosus lymphocytes. Hum. Mol. Genet. 29 2523–2534. 10.1093/hmg/ddaa131 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu J., Gao Y., Zhang Z., Zhang H. (2019f). Long noncoding RNA XIST acts as a competing endogenous RNA to promote malignant melanoma growth and metastasis by sponging miR-217. Panminerva Med. 7 63901–63912. 10.18632/oncotarget.11564 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhang H., Zhang W., Zhang Y., Wang W., Nie L. (2020d). LncRNA XIST modulates 5-hydroxytrytophan-induced visceral hypersensitivity by epigenetic silencing of the SERT gene in mice with diarrhea-predominant IBS. Cell. Signal. 73 109674–109683. 10.1016/j.cellsig.2020.109674 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhu Y., Gao G., Zhou Z. (2019g). Knockdown XIST alleviates LPS-induced WI-38 cell apoptosis and inflammation injury via targeting miR-370-3p/TLR4 in acute pneumonia. Cell Biochem. Funct. 37 348–358. 10.1002/cbf.3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. L., Li X. B., Hou Y. X., Fang N. Z., You J. C., Zhou Q. H. (2017). The lncRNA XIST exhibits oncogenic properties via regulation of miR-449a and Bcl-2 in human non-small cell lung cancer. Acta Pharmacol. Sin. 38 371–381. 10.1038/aps.2016.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. X., Zhu Z. B., Huang S. Z., Zhao Q., Huang C. J., Tang Y. H., et al. (2019h). lncRNA XIST regulates proliferation and migration of hepatocellular carcinoma cells by acting as miR-497-5p molecular sponge and targeting PDCD4. Cancer Cell Int. 19 198–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C. H., Bai X. F., Hu X. H. (2020). Knockdown of lncRNA XIST inhibits hypoxia-induced glycolysis, migration and invasion through regulating miR-381-3p/NEK5 axis in nasopharyngeal carcinoma. Eur. Rev. Med. Pharmacol. 24 2505–2517. [DOI] [PubMed] [Google Scholar]
- Zhao H., Wan J., Zhu Y. (2020). Carboplatin inhibits the progression of retinoblastoma through IncRNA XIST/miR-200a-3p/NRP1 axis. Drug Design Dev. Ther. 14 3417–3427. 10.2147/dddt.s256813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Zhao Y., He Y., Li Q., Mao Y. (2018). The functional pathway analysis and clinical significance of miR-20a and its related lncRNAs in breast cancer. Cell. Signal. 51 152–165. 10.1016/j.cellsig.2018.08.004 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Lu F., Su Q. L., Liu Z., Xia X. M., Yan Z. Z., et al. (2020). Knockdown of long noncoding RNA XIST mitigates the apoptosis and inflammatory injury of microglia cells after spinal cord injury through miR-27a/Smurf1 axis. Neurosci. Lett. 715 134649–134658. 10.1016/j.neulet.2019.134649 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Li S., Xia N., Shi Y., Zhao C. M. (2018). Effects of XIST/miR-137 axis on neuropathic pain by targeting TNFAIP1 in a rat model. J. Cell. Physiol. 233 4307–4316. 10.1002/jcp.26254 [DOI] [PubMed] [Google Scholar]
- Zheng C., Bai C., Sun Q., Zhang F., Yu Q., Zhao X., et al. (2020). Long noncoding RNA XIST regulates osteogenic differentiation of human bone marrow mesenchymal stem cells by targeting miR-9-5p. Mech. Dev. 162 103612–103621. 10.1016/j.mod.2020.103612 [DOI] [PubMed] [Google Scholar]
- Zheng R., Lin S., Guan L., Yuan H., Liu K., Liu C., et al. (2018). Long non-coding RNA XIST inhibited breast cancer cell growth, migration, and invasion via miR-155/CDX1 axis. Biochem. Biophys. Res. Commun. 498 1002–1008. 10.1016/j.bbrc.2018.03.104 [DOI] [PubMed] [Google Scholar]
- Zheng W., Li J., Zhou X., Cui L., Wang Y. (2020). The lncRNA XIST promotes proliferation, migration and invasion of gastric cancer cells by targeting miR-337. Arab J. Gastroenterol. 21 199–206. 10.1016/j.ajg.2020.07.010 [DOI] [PubMed] [Google Scholar]
- Zhou H., Sun L. N., Wan F. S. (2019a). Molecular mechanisms of TUG1 in the proliferation, apoptosis, migration and invasion of cancer cells. Oncol. Lett. 18 4393–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H. Q., Xian W., Zhang Y. X., Chen G., Zhao S., Chen X., et al. (2018). Trends in incidence and associated risk factors of suicide mortality in patients with non-small cell lung cancer. Cancer Med. 7 4146–4155. 10.1002/cam4.1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li D., Yang B. P., Cui W. J. (2020). LncRNA XIST inhibits hypoxia-induced cardiomyocyte apoptosis via mediating miR-150-5p/Bax in acute myocardial infarction. Eur. Rev. Med. Pharmacol. 24 1357–1366. [DOI] [PubMed] [Google Scholar]
- Zhou K., Li S., Du G., Fan Y., Wu P., Sun H., et al. (2019b). LncRNA XIST depletion prevents cancer progression in invasive pituitary neuroendocrine tumor by inhibiting bFGF via upregulation of microRNA-424-5p. OncoTargets Ther. 12 7095–7109. 10.2147/ott.s208329 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhou K., Yang J., Li X., Chen W. (2019c). Long non-coding RNA XIST promotes cell proliferation and migration through targeting miR-133a in bladder cancer. Exp. Ther. Med. 18 3475–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Qin G., Yang L., Xiang D., Li S. (2019d). LncRNA XIST regulates myocardial infarction by targeting miR-130a-3p. J. Cell. Physiol. 234 8659–8667. 10.1002/jcp.26327 [DOI] [PubMed] [Google Scholar]
- Zhou X. Y., Xu X. H., Gao C., Cui Y. S. (2019e). XIST promote the proliferation and migration of non-small cell lung cancer cells via sponging miR-16 and regulating CDK8 expression. Am. J. Transl. Res. 11 6196–6206. [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Zheng T., Yu J., Zhou L. X., Wang L. (2018). LncRNA XIST accelerates cervical cancer progression via upregulating Fus through competitively binding with miR-200a. Biomed. Pharmacother. 105 789–797. 10.1016/j.biopha.2018.05.053 [DOI] [PubMed] [Google Scholar]
- Zhu J., Zhang R., Yang D., Li J., Yan X., Jin K., et al. (2018). Knockdown of long non-coding RNA XIST inhibited doxorubicin resistance in colorectal cancer by upregulation of miR-124 and downregulation of SGK1. Cell. Physiol. Biochem. 51 113–128. 10.1159/000495168 [DOI] [PubMed] [Google Scholar]
- Zhuang L. K., Yang Y. T., Ma X., Han B., Wang Z. S., Zhao Q. Y., et al. (2016). MicroRNA-92b promotes hepatocellular carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST. Cell Death Dis. 7:e2203. 10.1038/cddis.2016.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Chen F. R., Xia R. P., Wang H. W., Xie Z. R., Xu Y., et al. (2020). Long noncoding RNA XIST regulates the EGF receptor to promote TGF-beta1-induced epithelial-mesenchymal transition in pancreatic cancer. Biochem. Cell Biol. 98 267–276. 10.1139/bcb-2018-0274 [DOI] [PubMed] [Google Scholar]
- Zuo K., Zhao Y., Zheng Y., Chen X. L., Du S., Liu Q. (2019). Long non-coding RNA XIST promotes malignant behavior of epithelial ovarian cancer. OncoTargets Ther. 12 7261–7267. 10.2147/ott.s204369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylicz J. J., Bousard A., Zumer K., Dossin F., Mohammad E., da Rocha S. T., et al. (2019). The implication of early chromatin changes in X chromosome inactivation. Cell 176 182–197. 10.1016/j.cell.2018.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]