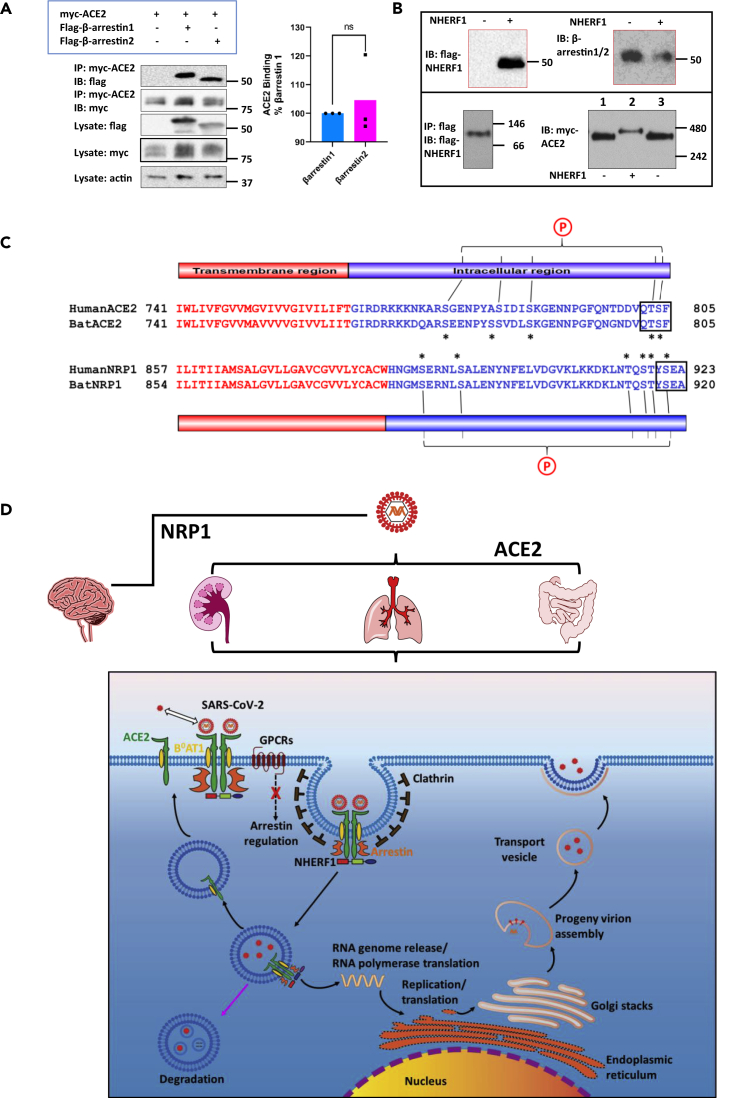
Figure 4.
Proposed SARS-CoV-2 cell entry mechanism regulated by PDZ-mediated interaction
(A) β-Arrestin 1 or β-arrestin 2 binds to ACE2 with nearly equivalent affinity. FLAG-β-arrestin 1 or 2 was transiently coexpressed in HEK293 GnTI cells with Myc-ACE2. The cleared cell lysates were incubated with anti-c-Myc agarose beads. Proteins are eluted by 2x Laemmli sample buffer (Bio-Rad) supplemented with β-mercaptoethanol and detected by western blotting using the indicated specific antibodies (left panel). β-Actin was used as a loading control. The molecular markers are indicated in kDa on the right. Given the equivalent expression of myc-ACE2 observed in the cell lysate and immunoprecipitates, pull-down of β-arrestin 1 or β-arrestin 2 by myc-ACE2 was quantified by normalizing the signal intensities to their individual lysate expression, respectively (right panel). Error bars indicate SD.
(B) NHERF1 is not required to promote ACE2 dimerization. FLAG-NHERF1 and Myc-ACE2 were transiently transfected into HK2 cells. Forty-eight hours post transfection, the cell lysates were cleared by centrifugation and incubated with anti-c-Myc agarose beads (only the sample in the lower left panel was prepared using anti-FLAG agarose beads and therefore eluted by 3x FLAG peptide). Purified proteins by c-Myc peptide elution were detected by the indicated antibodies. The boxed blots in red are from SDS-PAGE (top panel) and the blots in the lower panel are from blue native PAGE performed as described before (Dewson, 2015). Molecular weights are indicated at the right side of the blots. In the lower right panel, the same samples were loaded in the first and third lane to better show the slight difference of protein size from different samples.
(C) Sequence alignment of ACE2 and NRP1 from SARS-CoV-2 hosts human (Protein ID: NP_001358344.1 for ACE2 and ADN93470.1 for NRP1) and bat (Protein ID: NP_001231902.2 for ACE2 and KAF6372190.1 for NRP1). Human and bat ACE2/Ace2 and NRP1/Nrp1 are used because they are primary hosts of SARS-CoV-2 (Burki, 2020). Both identified SARS-CoV-2 receptors are single-pass transmembrane proteins and have a typical type I PDZ-binding motif (QTSF and YSEA) implying a possible common mechanism by which NHERF1 regulates SARS-CoV-2 receptor internalization. The single-pass transmembrane regions are colored in red, and the intracellular C-terminal tail is colored in blue. The conserved serine and threonine residues are indicated by asterisks and are likely to be potential phosphorylation sites denoted by an encircled P. These sites may mediate SARS-CoV-2 receptor binding to β-arrestin as in the case of β2AR, where the phosphorylated receptor C terminus is required for binding to β-arrestin and receptor internalization (Nobles et al., 2011).
(D) NHERF1 functions in ACE2-mediated SARS-CoV-2 cell entry in lung, kidney, and intestine and possibly NRP1-mediated virus entry in brain. NHERF1 enhances plasma membrane localization of B0AT1-stabilized ACE2 receptor and regulates its internalization and hence, ACE2-mediated SARS-CoV-2 cell entry. Except NHERF1, the virion-receptor complex also requires β-arrestin to facilitate its internalization process in which clathrin is involved. Once internalized, SAS-CoV-2 is either degraded in lysosomes (pink arrow) or released into the cytosol where the viral genomic RNA is released and immediately translated into viral RNA polymerase. The ACE2 receptor may be recycled back to the plasma membrane. Viral RNA genome and its structural nucleocapsid protein is replicated and synthesized in the cytoplasm. Other viral structural proteins are translated in the endoplasmic reticulum and further glycosylated in the Golgi. Mature progeny virions are then formed and bud out from plasma by exocytosis.