Highlights
-
•
The MinION sequencer allows whole HPV genome sequencing.
-
•
The proposed bioinformatics pipeline enables the reconstruction of whole HPV genome.
-
•
Rapid whole-genome characterization of HPV genomes with a minimal error rate.
Abbreviations: HPV, human papillomavirus; RCA, rolling circle amplification; NGS, next-generation sequencing; ONT, Oxford Nanopore Technologies
Keywords: MinION sequencer, Oxford Nanopore Technologies, Whole-genome sequencing, Papillomavirus
Abstract
Background
The MinION sequencer belongs to the third generation of sequencing technology that allows for the generation of ultra-long reads, representing a potentially more effective approach to characterize entire viral genome sequences than other time-consuming and low-throughput methodologies.
Methods
We report the use of the MinION nanopore sequencer to sequence the full-length genome of human papillomavirus (HPV)-ICB2 (7441 bp), which was previously characterized in our laboratory. Three independent MinION libraries were prepared and sequenced using either three consecutive 12 -h runs (Protocol A) or a single run of 48 h starting from a pool of three barcoded DNA libraries (Protocol B). A fully automated bioinformatics pipeline was developed for the reconstruction of the viral genome.
Results
Protocols A and B generated 9,354,933 and 3,255,879 reads, respectively. Read length N50 values ranged between 6976 and 7360 nucleotides over the four sequencing runs. Bioinformatics analysis showed that both protocols allowed for the reconstruction of the whole viral genome, with pairwise percentages of identity to HPV-ICB2 of 100 % for protocol A and 99.98 % for protocol B.
Conclusion
Our results show that the use of the MinION nanopore sequencer represents an effective strategy for whole-genome sequencing of HPVs with a minimal error rate.
1. Introduction
Human papillomaviruses (HPVs) are non-enveloped, icosahedral, double-stranded circular DNA viruses of approximately 8 kb in size [reviewed in (Gheit, 2019)]. The L1 gene that encodes the major capsid protein is the most conserved region in the PV genome; thus, it is used for the classification of these viruses [reviewed in (Bernard et al., 2010; de Villiers, 2013; de Villiers et al., 2004)]. PVs are classified into 53 different genera, among which alpha, beta, gamma, mu, and nu, that include human mucosal and cutaneous PVs. At present, 222 HPVs are listed by the International HPV Reference Center (www.hpvcenter.se) (Bernard et al., 2010; de Villiers, 2013; Gheit, 2019; Mühr et al., 2018; Tommasino, 2014). A subgroup of the genus Alphapapillomavirus, referred to as mucosal high-risk HPV types, are associated with anogenital cancer and a subset of head and neck cancers (Bouvard et al., 2009; Tommasino, 2014). The genus beta includes more than 50 beta HPV types divided into 5 species (de Villiers, 2013). Beta HPV types are abundantly present in the skin, and together with ultraviolet radiation, appear to be associated with the development of cutaneous squamous cell carcinoma (Orth, 2006; Tommasino, 2017). The gamma genus includes more than 100 HPV types divided into 27 species, and this number continues to expand (Brancaccio et al., 2018, 2017; Dutta et al., 2018, 2017). However, their potential role in human diseases remains unclear.
The recent advent of new molecular tools such as next-generation sequencing (NGS) has accelerated the discovery of new HPVs. In our laboratory, we developed an effective protocol for the isolation and characterization of new HPV types that combines the use of specific and degenerate primers targeting the L1 gene and NGS technology (Brancaccio et al., 2018). This strategy allowed us to obtain partial L1 sequences from putative new HPV types. Two studies have confirmed the validity of this protocol for the isolation of full viral genomes (Brancaccio et al., 2019, 2017). In these studies, the use of rolling circle amplification (RCA) to enrich circular DNA followed by long-range PCR using outward-directed specific PCR primers allowed the whole HPV genomes of HPV224 (species beta-2) and HPV-ICB2 (species gamma-8) to be obtained (Brancaccio et al., 2019, 2017). After a cloning step, the sequences of the complete viral genomes were obtained by Sanger sequencing in duplicate, which was laborious and time-consuming (Brancaccio et al., 2018).
To circumvent these limitations, we have evaluated the MinION nanopore sequencer, which allows for the generation of reads that are several kilobases (kb) in length, as a possible alternative strategy for sequencing the whole viral genome. This technology has previously been coupled to RCA for discovery of new episomal circular viruses (Vanmechelen et al., 2017b). The MinION belongs to the third generation of sequencing approaches and uses a nanopore-based method that is able to produce long reads of up to hundreds of thousands of bases (Goldstein et al., 2019; Jansen et al., 2017; Pennisi, 2012; Reiner et al., 2012). Moreover, the sequencer is portable, is small in size, and provides data in real time, allowing for rapid sequencing in the field or in clinical settings (McNaughton et al., 2019). Despite the constant improvement and development of new pipelines for data analysis, the primary limitations of this technology remain the error rate and accuracy of the raw reads, which is not yet comparable to NGS technologies (Kasianowicz et al., 1996; Keller et al., 2018; Lu et al., 2016; Makałowski and Shabardina, 2019; McNaughton et al., 2019). However, by performing a viral DNA enrichment step using RCA or PCR, the sequencing depth can be increased, thus influencing the attainable consensus accuracy (Li et al., 2016; McNaughton et al., 2019). Several studies have shown that reconstruction of entire genomes of specific viruses can be achieved with third-generation sequencing portable technology (Batovska et al., 2017; Wawina-Bokalanga et al., 2019).
This study aimed to determine whether the MinION sequencer can be used to obtain the whole genome of a previously characterized HPV (HPV-ICB2) with a minimal error rate, thus constituting a valid approach for the whole characterization of any papillomavirus.
2. Methods
2.1. Human specimen
HPV-ICB2 (accession number MK080568), a novel beta-2 HPV that is 7441 bp in length, was previously isolated from a human forearm skin swab sample and fully characterized in our laboratory (Brancaccio et al., 2019). This sample was collected from a patient enrolled in the VIRUSCAN study, a five-year (2014–2019) prospective cohort study conducted at the Moffitt Cancer Center and the University of South Florida (R01CA177586-01; “Prospective study of cutaneous viral infections and non-melanoma skin cancer”). The study was approved by the University of South Florida Institutional Review Board, and all participants provided written informed consent. All methods were carried out in accordance with relevant guidelines and regulations.
2.2. Rolling circle amplification
DNA was extracted and purified from a HPV-ICB2-positive skin swab as described previously (Schowalter et al., 2010). The DNA was amplified by multiply-primed rolling circle amplification (RCA) with random hexamer primers using an Illustra TempliPhi 100 Amplification kit following the manufacturer’s recommendations (GE Healthcare, Piscataway, NJ), with supplementation of 450 μM dNTPs as described by Rector et al. (2004).
2.3. MinION library preparation
Three libraries were prepared independently following the SQK-LSK109 protocol for 1D PCR barcoding amplicons (Fig. 1). First, long-range PCR was performed to amplify the entire HPV-ICB2 genome from a 1:100 diluted RCA product using a KAPA HIFI HotStart ReadyMixPCR kit following the manufacturer’s instructions (KAPA Biosystems, Boston, MA, USA) with HPV-ICB2-specific outward-directed primers tailed with the universal sequences provided by the manufacturer. The following tailed primers were used at a final concentration of 1.6 pM each: forward primer, 5′-TTT CTG TTG GTG CTG ATA TTG CCA GAC AGA ACA CAT CTT TTG ATC C-3′ and reverse primer, 5′-ACT TGC CTG TCG CTC TAT CTT CTC GTC CCG TGA CCC ACC CTG A-3′. All the above-mentioned primers were designed within a partial L1 region sequence of HPV-ICB2 (99 bp) obtained from previous next-generation sequencing data (Brancaccio et al., 2019, 2018). All the amplification steps were performed using proofreading polymerases.
Fig. 1.
Schematic overview of the research strategy for the full-length viral genome characterization of papillomaviruses. Partial L1 sequences from putative new HPV types are available in GenBank or from next-generation sequencing data. After identification by PCR of HPV DNA-positive samples for the corresponding L1 sequence, multiply primed rolling circle amplification (RCA) was performed on the corresponding DNA sample to enrich circular DNA. To obtain the complete viral genome, first, long-range PCR was performed to amplify the entire HPV genome from a 1:100 diluted RCA product, using specific outward-directed primers tailed with the Oxford Nanopore universal tags. A second PCR to add the barcodes from the barcoding kit (EXP-PBC001) was performed. After DNA repair, end-prep, adapter ligation, and clean-up steps, the libraries were loaded into the MinION flow cell consecutively (Protocol A: three separate 12 -h runs) or simultaneously (Protocol B: a single 48 -h run).
PCR amplification was performed in a C1000 Touch thermal cycler (Bio-Rad Laboratories, Inc.) using the following thermocycling conditions: an initial denaturation step of 3 min at 95 °C followed by 35 cycles of denaturation at 98 °C (20 s), annealing at 64 °C (15 s), and extension at 72 °C (8.5 min), with a final extension at 72 °C (10 min) to generate a 7485 bp product. The amplicon was then purified using a QIAquick gel extraction kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions.
One barcode was added to each of the three purified amplicons by performing a second PCR step using a KAPA HIFI HotStart ReadyMixPCR kit (KAPA Biosystems, Boston, MA, USA). Three barcodes (i.e., BC01, BC02, and BC03 for runs 1, 2, and 3, respectively) provided in the PCR Barcoding Expansion 1–12 kit (EXP-PBC001, Oxford Nanopore Technologies, Oxford, UK) were used.
PCR amplification was performed using the following thermocycling conditions: an initial denaturation step of 3 min at 95 °C was followed by 35 cycles of denaturation at 98 °C (20 s), annealing at 62 °C (15 s), and extension at 72 °C (8.5 min), with a final extension at 72 °C for 8.5 min. The resulting amplicon was then purified using a 0.6× ratio of Agencourt AMPure XP beads (Beckman Coulter, Pasadena, California, USA) following the manufacturer’s instructions and then eluted in 40 μL of Invitrogen UltraPure DNase/RNase-Free distilled water (Gibco, Life Technologies, Paisley, UK).
2.4. MinION sequencing run
DNA repair, end-prep, adapter ligation, and clean-up steps were performed following the SQK-LSK109 protocol (Oxford Nanopore Technologies, Oxford, UK). The final product was quantified using a Qubit fluorometer (Thermo Fisher Scientific, Waltham, USA) and loaded into the MinION flow cell, with sequencing performed according to the manufacturer’s instructions (Oxford Nanopore Technologies, Oxford, UK) using a FLO-MIN106D flow cell. Two sequencing protocols (A and B) were established. In protocol A, three separate 12 -h runs (referred to as runs 1, 2, and 3) were performed using the same flow cell. The libraries BC1, BC02, and BC03 were sequenced independently. Protocol B involved a single run of 48 h (referred to as run 4) using the remaining active nanopores and was performed with a pool of the three libraries (BC01, BC02, and BC03). After each run, the flow cell was washed using a Flowcell Wash Kit (EXP-WSH002) from Oxford Nanopore Technologies, following the manufacturer’s instructions.
All the sequencing runs were performed applying the default starting voltage of −180 mV.
2.5. Bioinformatics data analysis
Coverage was calculated using SAMtools, and the percentage of similarity was determined by the number of matches or mismatches at each base using Pysamstats v1.1.2 (https://github.com/alimanfoo/pysamstats) (Batovska et al., 2017). Both coverage and similarity were computed on the filtered reads obtained after two steps of quality and size filtering using NanoFilt and Filtlong tools. The fully automated bioinformatics pipeline is described in Fig. 2 (https://github.com/IARCbioinfo/MinION_pipes). A Supplementary file S1 describing all the bioinformatics tools and parameters is also available. Guppy v4.0.15+ (https://community.nanoporetech.com/protocols/Guppy-protocol) (Adrien Leger and Tommaso Leonardi, 2019; Wick et al., 2019) was used to perform base-calling, de-multiplexing and barcode trimming steps. NanoFilt v2.7.1 (https://github.com/wdecoster/nanofilt) (De Coster et al., 2018) was used to remove unspecific reads longer than 8000 bp. Filtlong v0.2.0 (https://github.com/rrwick/Filtlong) was used to remove reads shorter than 7000 bp and low-quality reads (the worst 90 % of the reads were removed). To assess the effectiveness of the filtering steps, defined as the similarity between the draft sequence and the reference, two BLAST analyses, using Megablast algorithm, were performed on the raw reads and on the filtered reads. Canu v2.1 was used to reconstruct the whole genome of HPV-ICB2. Medaka v1.0.3 (https://github.com/nanoporetech/medaka) (“Medaka: Sequence correction provided by ONT Research (Accessed 27 Jun 2020).,” n.d.) was used for the polishing of the draft genomes. The filtered reads, generated by Filtlong were used as reference for this polishing step.
Fig. 2.
Pipeline for the processing of MinION data. Guppy v4.0.15+ (https://community.nanoporetech.com/protocols/Guppy-protocol) was used to perform base-calling, de-multiplexing and barcode trimming steps. NanoFilt v2.7.1 was used to remove unspecific reads longer than 8000 bp. Filtlong v0.2.0 was used to remove reads shorter than 7000 bp. The worst 90 % of the reads were removed. Canu v2.1 was used to reconstruct the whole genome of HPV-ICB2. Medaka v1.0.3 was used for the polishing of the draft genomes.
Finally, a consensus sequence was obtained by aligning the genomes generated from the three sequencing runs using MUSCLE (Edgar, 2004).
2.6. Availability of workflows, tools, and code
The raw sequencing data are available in the Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra), under BioProject PRJNA673786. All the code can be retrieved on the IARC bioinformatics platform on GitHub (https://github.com/IARCbioinfo/MinION_pipes).
3. Results
3.1. MinION sequencing and assembly using three independent runs (Protocol A)
Three consecutive 12 -h sequencing runs, each using an independent DNA library, were performed with the 1D PCR barcoding amplicons (SQK-LSK109) protocol on a MinION flow cell (R9.4.1). Before each sequencing run, the number of available nanopores was measured using the protocol “check your flow cell” in the MinKNOW software. The number of active nanopores decreased from run 1 to run 3 (1577, 1124, and 857 respectively), thus exceeding the guaranteed level (800 pores). The combined runs generated 9,354,933 raw reads (run 1: 3,186,245 reads; run 2: 2,464,705 reads; and run 3: 3,703,983 reads). More than 93 % of the reads generated by runs 1, 2 and 3 passed the quality control (QC) filtering (Table 1).
Table 1.
Sequencing output metrics and sequencing quality data using the ONT 1D kit (SQK-LSK109) from three MinION runs (Protocol A). A fourth run was performed using the remaining active nanopores (N = 560) with a pooled library (Protocol B).
| Protocol | Run | Active pores (N) | Running time (hrs) | Barcodes used | Reads with barcode (%) | Raw reads (N) | Gigabases called (Gb) | QC filtered reads (%) | Mean read length (nt) | N50 (nt) | Mean ICB2 coverage (fold) | Mean similarity to HPV-ICB2 (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 1 | 1577 | 12 | BC01 | 92.3 | 3,186,245 | 6.45 | 94.7 | 2025 | 7327 | 68,706 | 97.48 |
| 2 | 1124 | 12 | BC02 | 93.5 | 2,464,705 | 5.58 | 94.9 | 2266 | 7360 | 56,969 | 98.03 | |
| 3 | 857 | 12 | BC03 | 94.4 | 3,703,983 | 3.97 | 93.0 | 1070 | 7303 | 37,635 | 97.83 | |
| B | 4 | 560 | 48 | BC01/02/03 | 88.5 | 3,255,879 | 3.96 | 88.2 | 1253 | 6976 | 13,310 | 97.60 |
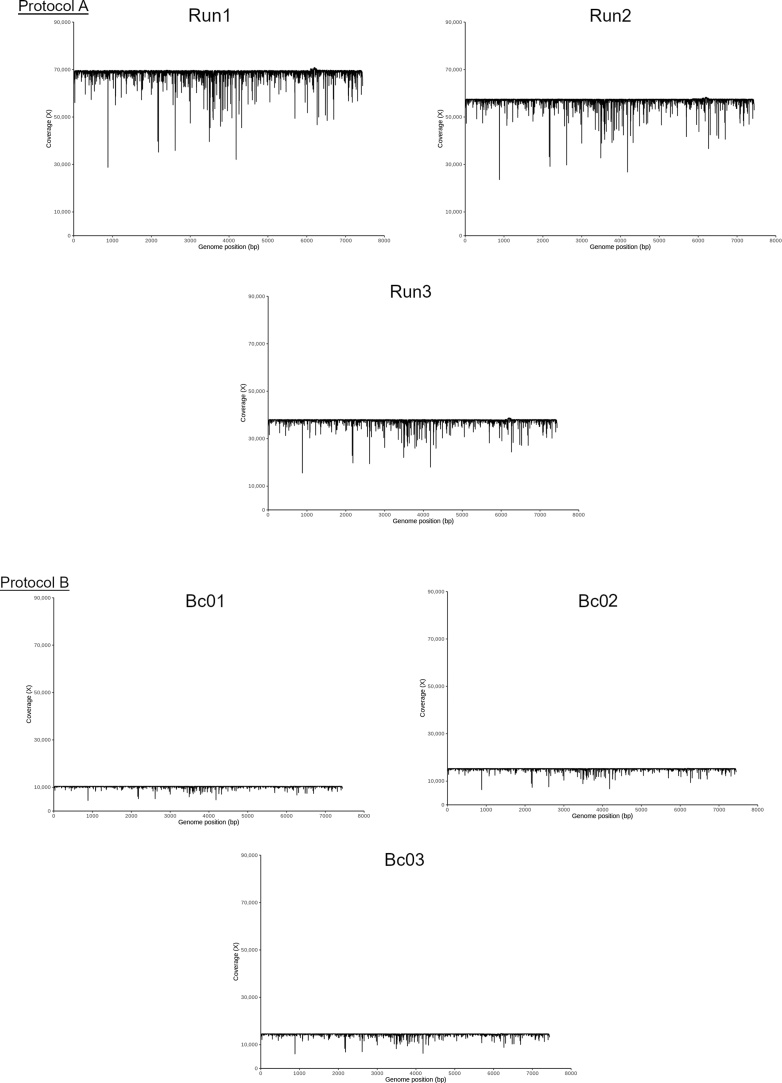
The N50 value, which describes the sequence length where 50 % of the sequenced bases are contained within reads of this length or longer, was greater than 7300 nt for all three runs (Table 1). Fig. 3A shows the total yield in gigabases generated over time for each of the sequencing runs, and Fig. 3B shows the number of reads and their length. The proportion of reads that failed the QC filtering step increased from run 1 (5.3 %) to run 3 (7.0 %), and the coverage of the HPV-ICB2 genome was higher in run 1 (mean coverage: 68,706-fold) than in runs 2 and 3, which had coverages of 56,969-fold and 37,635-fold, respectively (Table 1 and Fig. 4A).
Fig. 3.
(A) Total yield in gigabases generated over time for each of the sequencing runs. Three 12 -h runs (runs 1–3; Protocol A) and a 48 -h single run (run 4) with pooled libraries (Protocol B) were performed using a FLO-MIN106D flow cell. The blue line represents all the reads, while the green line represents only the reads with a Q ≥ 7.(B) Number of reads and their length in protocol A (runs 1–3) and protocol B (run 4). The graphs were generated using MinIONQC(Lanfear et al., 2019) (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
Fig. 4.
(A) Coverage of the MinION reads when aligned to the reference HPV-ICB2 genome for runs 1–3 (protocol A). (B) Coverage of the MinION reads when aligned to the reference HPV-ICB2 genome for run 4 (Protocol B).
Similarity analysis of MinION reads to the reference genome HPV-ICB2 was performed (Fig. 5A), and the results showed the presence of seven nucleotide positions (positions 882, 2184, 2612, 3001, 3301, 3491 and 3592) with the highest error rate in all three runs. The analysis of the raw data showed that the average error rates were 2.52 %, 1.97 %, and 2.17 % for runs 1, 2, and 3, respectively (Table 1).
Fig. 5.
(A) Percentage of similarity among each base when aligned to the HPV-ICB2 reference genome for runs 1–3 (Protocol A). (B) Percentage of similarity among each base when aligned to the HPV-ICB2 reference genome for run 4 (Protocol B) (B).
The reads were identified using the BLAST tool and the whole nucleotide collection (nr/nt, October 2020) database. The majority of the reads were representative of HPV-ICB2 (77.5 %, 92.3 %, and 63.2 % for runs 1, 2, and 3, respectively) while the remaining reads were representative of bacteria, archaea, eukaryota, and other viral sequences (Supplementary Table S1).
A second BLAST alignment was performed after read-filtering (Fig. 2), which allowed for 99.9 % of the sequences identified as HPV-ICB2 to be obtained in all three runs (Supplementary Table S1).
The whole genome of HPV-ICB2 was then obtained using Canu. The percentage of identity to HPV-ICB2 was 99.95 %, 99.89 %, and 99.97 % for runs 1, 2, and 3, respectively (data not shown). Subsequently, the polishing step using Medaka improved the percentage of identity to HPV-ICB2, reaching 100 %, 99.98 %, and 100 % for runs 1, 2, and 3, respectively (Table 2 and Fig. 2).
Table 2.
Effect of run time on the sequencing output metrics and identity of the reconstructed sequences against the reference genome of HPV-ICB2.
| Protocol | Run | Library ID | Running time (hrs) | Mean HPV-ICB2 coverage (fold) | Mean similarity to HPV-ICB2 (%) | Barcoded reads (N) | N50 (nt) | Mean read length (nt) | Identity to HPV-ICB2 (%) | Error type | Errors (nt position) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 1 | BC01 | 3h | 17446 | 97.48 | 863729 | 7335 | 1623 | 99.98 | 1 gap | 3571 |
| 6h | 35345 | 97.48 | 1550247 | 7339 | 1823 | 99.98 | 1 gap | 3571 | |||
| 9h | 52363 | 97.47 | 2190304 | 7340 | 1908 | 100 | — | — | |||
| 12 h (end of the run) | 68706 | 97.48 | 2808901 | 7339 | 1949 | 100 | — | — | |||
| 2 | BC02 | 3h | 14730 | 98.12 | 626541 | 7365 | 1854 | 99.98 | 1 gap | 3571 | |
| 6h | 29870 | 98.07 | 1135236 | 7373 | 2073 | 99.98 | 1 gap | 3511 | |||
| 9h | 44036 | 98.05 | 1585583 | 7375 | 2186 | 100 | — | — | |||
| 12 h (end of the run) | 56969 | 98.03 | 1991740 | 7376 | 2249 | 99.98 | 1 gap | 3571 | |||
| 3 | BC03 | 3h | 10514 | 97.99 | 1085013 | 7354 | 784 | 100 | — | — | |
| 6h | 21135 | 97.90 | 1910674 | 7367 | 893 | 99.97 | 2 gaps | 3511, 3580 | |||
| 9h | 30285 | 97.88 | 2577642 | 7371 | 947 | 100 | — | — | |||
| 12 h (end of the run) | 37635 | 97.83 | 3112798 | 7373 | 974 | 100 | — | — | |||
| B | 4 | BC01 | 3h | 2296 | 97.93 | 146863 | 7360 | 1268 | 100 | — | — |
| 6h | 4800 | 97.86 | 273473 | 7365 | 1417 | 100 | — | — | |||
| 9h | 6847 | 97.81 | 366143 | 7370 | 1505 | 99.98 | 1 gap | 3571 | |||
| 12h | 8414 | 97.76 | 438337 | 7368 | 1542 | 100 | — | — | |||
| 48 h (end of the run) | 10325 | 97.68 | 594410 | 7338 | 1387 | 99.98 | 1 gap | 3490 | |||
| BC02 | 3h | 3433 | 97.93 | 152174 | 7348 | 1786 | 99.95 | 3 gaps | 3591, 6019, 6080 | ||
| 6h | 7108 | 97.81 | 285539 | 7357 | 1964 | 100 | — | — | |||
| 9h | 10089 | 97.85 | 385430 | 7364 | 2066 | 99.98 | 1 gap | 3571 | |||
| 12h | 12376 | 97.77 | 463636 | 7363 | 2104 | 99.95 | 3 gaps | 3580, 3584, 3591 | |||
| 48 h (end of the run) | 15165 | 97.67 | 628925 | 7330 | 1893 | 99.98 | 1 gap | 3511 | |||
| BC03 | 3h | 4483 | 97.64 | 436785 | 7227 | 822 | 100 | — | — | ||
| 6h | 7897 | 97.62 | 736160 | 7252 | 856 | 100 | — | — | |||
| 9h | 10406 | 97.60 | 936322 | 7270 | 885 | 100 | — | — | |||
| 12h | 12232 | 97.56 | 1084010 | 7258 | 897 | 99.98 | 1 gap | 3571 | |||
| 48 h (end of the run) | 14441 | 97.45 | 1391886 | 7076 | 822 | 100 | — | — |
Finally, a consensus was obtained after the alignment of the three genomes using MUSCLE. The final percentage of identity to HPV-ICB2 was 100 %.
Of note, as reported elsewhere (Greninger et al., 2015; Tyler et al., 2018), we observed the presence of 211,436 BC1 reads in run 2 (9.0 % of the barcoded reads) and 33,959 BC1 and 142,887 BC2 reads in run 3 (1.0 % and 4.1 % of the barcoded reads respectively) due to carry-over DNA from previous runs, despite intermediate washing steps.
3.2. MinION sequencing and assembly using a single run (Protocol B)
A final 48 -h run (run 4) was performed using the remaining 560 active nanopores of the flow cell with a pool consisting of the three barcoded DNA libraries until the nanopores were exhausted. This run produced 3,255,879 reads, among which 88.2 % passed the QC filtering, and an N50 value of 6976 bp was obtained. More than 77 % of the barcoded reads generated by run 4 were identified as HPV-ICB2 sequences using BLAST, and a mean coverage of 13,310-fold was obtained (Supplementary Table S2 and Fig. 4B). Similarity analysis of MinION reads to the reference HPV-ICB2 genome allowed the identification of seven nucleotide positions (882, 2184, 2612, 3001, 3491, 4182 and 6268) with the highest error rate for run 4 (Fig. 5B). After reads filtering, more than 99.98 % of the reads were identified as belonging to the HPV-ICB2 genome (Supplementary Table S2).
The whole HPV-ICB2 genome was obtained from each barcoded library using Canu and showed a percentage of identity to HPV-ICB2 of 99.95 %, 99.94 %, and 99.90 % for BC1, BC2, and BC3, respectively, which increased to 99.98 %, 99.98 % and 100 % after a Medaka polishing step (Table 2, and data not shown). Finally, a consensus sequence was obtained after the alignment of the three genomes using MUSCLE. The final percentage of identity to HPV-ICB2 was 99.98 % (1 nucleotide gap).
The single nucleotide gap was located within a homopolymeric G stretch at the nucleotide position 3495 (Table 2). Sanger sequencing performed on the DNA libraries loaded into the flow cell demonstrated the absence of gaps before the sequencing run.
3.3. Effect of run time on final assembly quality
MinION sequencing data were analysed based on cumulative run time to simulate the effect of run-length on final assembly quality. Reads generated in the first 3, 6, 9, and 12 h for protocol A and 3, 6, 9, 12 and 48 h for protocol B, were analysed. Three hours was determined as the shortest run time sufficient to assemble the whole viral genome for both protocols A and B. After 3 h only, the coverages ranged from 2296-fold to 17,446-fold, and the mean percentages of similarity ranged from 97.48 % to 98.12 %, thus allowing the assembly of the whole viral genome with the percentage of identity to HPV-ICB2 ≥ 99.95 % for both protocols (Table 2, and data not shown).
4. Discussion
This study aimed to evaluate the capability of the MinION nanopore sequencer produced by Oxford Nanopore Technologies (ONT) to sequence the whole genome of a papillomavirus, using a single flow cell. HPV-ICB2, a new 7441-bp beta-2 PV, previously isolated in our laboratory from a human-derived specimen and fully characterized by Sanger sequencing (Brancaccio et al., 2019), was here used as a reference genome. To date, the methods used to identify and fully characterize new PVs have been based on Sanger sequencing and NGS (Arroyo et al., 2013; Barzon et al., 2011; Brancaccio et al., 2018; Bzhalava et al., 2014; Ekström et al., 2011; Johansson et al., 2013; Kocjan et al., 2015), which are low-throughput and time-consuming methodologies, respectively. Also, whereas such technologies generate short reads of only up to a few hundred nucleotides, the use of metagenomics data for viral genome reconstruction can lead to the artefactual assembly of chimeric genomes. This risk is particularly elevated when several related HPVs are present in the sample (Besser et al., 2018; Beaulaurier et al., 2020; Kuiama Lewandowski et al., 2019; McNaughton et al., 2019; Pastrana et al., 2018). Therefore, ONT technology may be useful for rapid sequencing of full-length viral genomes from PVs.
Several recent studies have reported the use of this technology for the sequencing of viral genomes from hepatitis B virus (McNaughton et al., 2019), simian immunodeficiency virus (Augier et al., 2019), Ebola virus (Wawina-Bokalanga et al., 2019), hepatitis C virus (Uchida et al., 2019), herpesvirus (Karamitros et al., 2016, p. 1), poxvirus (Prazsák et al., 2018b), parapoxvirus (Günther et al., 2017), or Zika virus (de Jesus et al., 2019), with accuracies of up to 99 %. A novel species of papillomavirus in giraffe lesions was also identified using this technology (Vanmechelen et al., 2017a). Also, many studies have used long-read sequencing for transcriptomic studies of herpesvirus (Boldogkői et al., 2018b; Prazsák et al., 2018a; Tombácz et al., 2019, 2018a), poxvirus (Tombácz et al., 2018b), and baculovirus (Boldogkői et al., 2018a; Moldován et al., 2018), or viral genome detection or analysis of avipoxvirus (Croville et al., 2018), henipavirus (Yinda et al., 2019), and poxvirus (Sasani et al., 2018).
To obtain an accurate sequence of the whole HPV-ICB2 genome and fully explore the potential of the MinION flow cell, in this study we established two sequencing protocols (A and B), both of which were based on the sequencing of three barcoded libraries, either independently or pooled.
The use of an automated bioinformatics pipeline, developed in our laboratory, enabled the reconstruction of the whole genome of HPV-ICB2 from both protocols, with no errors for protocol A and only one single error for protocol B. An HPV isolate is considered to be a new type when its L1 gene shows variations greater than 10 % relative to the closest known type. Thus, even the single error observed in protocol B is acceptable for identifying known or new HPV types.
The high representation of unexpected short or long length reads can be explained by unspecific or incomplete amplifications during the barcoding step, environmental contaminants (e.g. bacteria), DNA degradation or ligation events, or incomplete sequencing. This result highlights the importance of performing size-based filtering steps to remove non-HPV-ICB2 reads. Before performing these steps, up to 36.8 % of the reads were classified as belonging to archaea, eukaryotes, or bacteria, whereas BLAST analysis performed after the filtering step showed that at least 99.96 % of the sequences were identified as belonging to the HPV-ICB2 reference genome. The identification after filtering steps of less than 0.042 % of non-HPV-ICB2 reads reflect the presence of environmental contaminants in the library preparation (i.e. bacteria, viruses), the presence of other HPVs that were unexpectedly amplified despite the use of HPV-ICB2 specific primers, or low quality reads that were misclassified. The raw MinION sequencing data were filtered using two different tools, NanoFilt and Filtlong, for removing reads of unexpected size (< 7000 bp and > 8000 bp). The latter filtering tool is based on “the final score”, a combination of three component scores: length score, mean quality score, and window quality score. Thus, these two filtering steps allowed the selection of high quality reads of the expected size. Moreover, Filtlong has the possibility of providing a reference sequence that can be taken into account in the filtering of reads. However, in this study, the whole genome of HPV-ICB2 was obtained without a reference genome thus confirming the effectiveness of our strategy for reconstructing unknown HPV genomes. After filtering, and the reconstruction of draft genomes using Canu, a final polishing step was performed, using Medaka. This step slightly improved the quality of the draft genomes.
The single error (1 nucleotide gap) in protocol B, after 48 h of sequencing run may represent a systematic error. Moreover, sequencing errors may also be introduced during the bioinformatics analysis (e.g., base-calling or polishing steps), as reported elsewhere (McNaughton et al., 2019). However, the use of other sequencing flow cells and chemistries (e.g., R10.3 chemistry), and the development of ONT technologies with higher accuracy may further reduce the error rate (Tyler et al., 2018). All the nucleotide gaps reported in this study were due to nucleotide insertion within homopolymeric stretches. This base-calling limitation of the Oxford Nanopore technology has been reported elsewhere (Tyler et al., 2018). However, most of the cases, the use of Canu allowed us to fix the systematic errors allowing the generation of whole HPV-ICB2 genomes with a perfect identity (100 %) to the reference genome of the virus. No PCR amplification artefacts were found in this study.
Despite the small number of active nanopores available (N = 560) for running protocol B, approximately 76 % of the barcoded reads were generated within 12 h, allowing for the reconstruction of the whole genome of HPV-ICB2 with a percentage of identity of 99.98 %. Thus, we assume that the use of optimal conditions (i.e., number of active nanopores > 800) and a single run of up to 48 h should allow for the generation of good-quality sequencing data for the reconstruction of up to four viral genomes in triplicate.
Moreover, our data showed that a sequencing run of 3 h was determined as the shortest run time sufficient to assemble the full-length viral genome from both protocols, with the percentage of identity to HPV-ICB2 ≥ 99.95 %.
All the sequencing runs were performed applying the default starting voltage of −180 mV. Especially for run 4, adjustments of the starting voltage could have been considered to obtain better results. However, the voltage is automatically controlled by the MinKNOW software for the rest of the run time, and thus we consider these adjustments to be of minor impact. The complete protocol described in this study can be performed in less than two weeks and represents an alternative to time-consuming methodologies for the whole-genome sequencing of HPVs; the RCA step can be performed in one day, followed by another day for the long-range PCR using HPV-ICB2-specific outward-directed primers, agarose gel purification, and library preparation. Then, the total duration of the protocol depends on the sequencing run times and on the complete bioinformatics analysis. The Guppy basecalling step was computationally expensive and time-consuming, thus constituting the longer part of our analysis.
As reported elsewhere (Greninger et al., 2015; Tyler et al., 2018), carry-over DNA contaminants from previous runs were detected despite intermediate washing steps. However, the data generated from runs 2 and 3 (Protocol A) showed that only 9.0 % and 5.1 % of the barcoded reads, respectively, were identified as contaminants from previous runs, and thus the impact of this technical limitation on our study is minimal. The use of different barcoded libraries on the same flow cells, when applying this protocol, is thus warranted. Most importantly, with the use of most recent ONT wash kits (e.g. EXP-WSH004), a flow cell should be able to be used for consecutive sequencing experiments with minimal carryover.
Xu et al. (2018) have recently reported another ONT limitation in multiplexing sequencing. The authors showed that less than 0.6 % of total reads were assigned to the incorrect barcode mainly by the generation of chimeric reads. This limitation could not be evaluated in our study as multiple sequencing of a single target has been performed. However, the steadily improvement of the Nanopore sequencing technology should greatly alleviate this limitation.
Also, we observed a progressive decrease in the number of active nanopores after each of the runs. We believe that together, the time spent running, possible storage of the flow cell between two runs, and the washing steps contribute to reducing the number of nanopores available for the sequencing.
Because too many nanopores become inactive after consecutive runs, a single multiplexed run with barcoded libraries might be considered in future protocols to obtain reads of sufficient quality and quantity to reconstruct different full-length PV genomes.
5. Conclusions
Our results showed that the use of the MinION nanopore sequencer is an effective and rapid strategy for whole-genome sequencing of HPVs. Furthermore, this technology can be easily integrated into our protocol for the isolation and full characterization of novel HPV genomes (Brancaccio et al., 2019, 2018, 2017).
The first part of our protocol for the discovery of novel papillomaviruses relies on the use of degenerate-primer PCR to amplify part of the viral genome, combined with next-generation sequencing (Brancaccio et al., 2018). This strategy led to the discovery of partial L1 sequences from 105 putative new PV types in the oral cavity and skin. Also, thousands of partial genomic sequences of putative novel HPV types have been deposited in GenBank and remain to be fully characterized, some of which may display transforming activity and deserve attention (reviewed in (Gheit, 2019)). Starting from a partial L1 region sequence (e.g., 99 bp for HPV-ICB2) and using long-range PCR using specific outward-directed primers with the RCA product as a template, the full-length genome of any papillomaviruses can potentially be obtained after MinION sequencing. We assume that using optimal conditions (i.e. number of active nanopores > 800), 12 unique barcodes, and a single run, the use MinION nanopore sequencer should allow the generation of good quality sequencing data for the reconstruction of up to 12 whole viral genomes, or up to four viral genomes in triplicate.
In summary, the protocol described here represents a valid alternative to classical sequencing strategies for the rapid and accurate whole-genome characterization of papillomaviruses.
Disclaimer
Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article, and they do not necessarily represent the decisions, policy, or views of the International Agency for Research on Cancer/World Health Organization.
Ethics approval and consent to participate
The sample was collected from a patient enrolled in the VIRUSCAN study, a five-year (2014-–2019) prospective cohort study conducted at the Moffitt Cancer Center and the University of South Florida (R01CA177586−01; “Prospective study of cutaneous viral infections and non-melanoma skin cancer”). The study was approved by the University of South Florida Institutional Review Board, and all participants provided written informed consent. All methods were carried out in accordance with relevant guidelines and regulations.
Availability of data and materials
The raw sequencing data are available in the Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra), under BioProject PRJNA673786. All the code can be retrieved on the IARC bioinformatics platform on GitHub (https://github.com/IARCbioinfo).
Funding
The study was partially supported by grants from the National Cancer Institute at the National Institutes of Health (R01-CA177586) to D.E.R. and from the Foundation ARC (PJA 20151203192) to M.T.
Authors contribution
All authors reviewed the manuscript.
CRediT authorship contribution statement
Rosario N. Brancaccio: Investigation, Software, Writing - original draft, Formal analysis, Validation, Data curation, Methodology, Writing - review & editing. Alexis Robitaille: Writing - review & editing. Sankhadeep Dutta: Resources. Dana E. Rollison: Resources, Funding acquisition, Writing - review & editing. Massimo Tommasino: Conceptualization, Supervision, Writing - original draft, Funding acquisition, Validation, Writing - review & editing. Tarik Gheit: Conceptualization, Investigation, Supervision, Writing - original draft, Formal analysis, Validation, Data curation, Methodology, Writing - review & editing.
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgments
We are grateful to Dr. Karen Müller for editing, and to Ms. Nicole Suty for her help with preparation of this manuscript.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jviromet.2021.114180.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Adrien Leger, Tommaso Leonardi pycoQC, interactive quality control for Oxford Nanopore Sequencing. Int. J. Open Source Softw. Process. 2019 doi: 10.21105/joss.01236. [DOI] [Google Scholar]
- Arroyo L.S., Smelov V., Bzhalava D., Eklund C., Hultin E., Dillner J. Next generation sequencing for human papillomavirus genotyping. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2013;58:437–442. doi: 10.1016/j.jcv.2013.07.013. [DOI] [PubMed] [Google Scholar]
- Augier C., Beyne E., Villabona-Arenas C.J., Mpoudi Ngole E., Peeters M., Ayouba A. Identification of a novel simian immunodeficiency virus-infected african green monkey (Chlorocebus tantalus) confirms that tantalus monkeys in Cameroon are infected with a mosaic SIVagm lineage. AIDS Res. Hum. Retroviruses. 2019 doi: 10.1089/AID.2019.0216. [DOI] [PubMed] [Google Scholar]
- Barzon L., Militello V., Lavezzo E., Franchin E., Peta E., Squarzon L., Trevisan M., Pagni S., Dal Bello F., Toppo S., Palù G. Human papillomavirus genotyping by 454 next generation sequencing technology. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2011;52:93–97. doi: 10.1016/j.jcv.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Batovska J., Lynch S.E., Rodoni B.C., Sawbridge T.I., Cogan N.O. Metagenomic arbovirus detection using MinION nanopore sequencing. J. Virol. Methods. 2017;249:79–84. doi: 10.1016/j.jviromet.2017.08.019. [DOI] [PubMed] [Google Scholar]
- Beaulaurier J., Luo E., Eppley J.M., Uyl P.D., Dai X., Burger A., Turner D.J., Pendelton M., Juul S., Harrington E., DeLong E.F. Assembly-free single-molecule sequencing recovers complete virus genomes from natural microbial communities. Genome Res. 2020;30:437–446. doi: 10.1101/gr.251686.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard H.-U., Burk R.D., Chen Z., van Doorslaer K., zur Hausen H., de Villiers E.-M. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401:70–79. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besser J., Carleton H.A., Gerner-Smidt P., Lindsey R.L., Trees E. Next-generation sequencing technologies and their application to the study and control of bacterial infections. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2018;24:335–341. doi: 10.1016/j.cmi.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogkői Z., Moldován N., Szűcs A., Tombácz D. Transcriptome-wide analysis of a baculovirus using nanopore sequencing. Sci. Data. 2018;5 doi: 10.1038/sdata.2018.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogkői Z., Szűcs A., Balázs Z., Sharon D., Snyder M., Tombácz D. Transcriptomic study of Herpes simplex virus type-1 using full-length sequencing techniques. Sci. Data. 2018;5 doi: 10.1038/sdata.2018.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvard V., Baan R., Straif K., Grosse Y., Secretan B., El Ghissassi F., Benbrahim-Tallaa L., Guha N., Freeman C., Galichet L., Cogliano V. WHO International Agency for Research on Cancer monograph Working Group, 2009. A review of human carcinogens--part B: biological agents. Lancet Oncol. 2009;10:321–322. doi: 10.1016/s1470-2045(09)70096-8. [DOI] [PubMed] [Google Scholar]
- Brancaccio R.N., Robitaille A., Dutta S., Rollison D.E., Fischer N., Grundhoff A., Tommasino M., Gheit T. Complete genome sequence of a novel human gammapapillomavirus isolated from skin. Genome Announc. 2017;5 doi: 10.1128/genomeA.00833-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancaccio R.N., Robitaille A., Dutta S., Cuenin C., Santare D., Skenders G., Leja M., Fischer N., Giuliano A.R., Rollison D.E., Grundhoff A., Tommasino M., Gheit T. Generation of a novel next-generation sequencing-based method for the isolation of new human papillomavirus types. Virology. 2018;520:1–10. doi: 10.1016/j.virol.2018.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancaccio R.N., Robitaille A., Dutta S., Rollison D.E., Tommasino M., Gheit T. Isolation of a novel Beta-2 human papillomavirus from skin. Microbiol. Resour. Announc. 2019;8 doi: 10.1128/MRA.01628-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzhalava D., Mühr L.S.A., Lagheden C., Ekström J., Forslund O., Dillner J., Hultin E. Deep sequencing extends the diversity of human papillomaviruses in human skin. Sci. Rep. 2014;4:5807. doi: 10.1038/srep05807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croville G., Le Loc’h G., Zanchetta C., Manno M., Camus-Bouclainville C., Klopp C., Delverdier M., Lucas M.-N., Donnadieu C., Delpont M., Guérin J.-L. Rapid whole-genome based typing and surveillance of avipoxviruses using nanopore sequencing. J. Virol. Methods. 2018;261:34–39. doi: 10.1016/j.jviromet.2018.08.003. [DOI] [PubMed] [Google Scholar]
- De Coster W., D’Hert S., Schultz D.T., Cruts M., Van Broeckhoven C. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics. 2018;34:2666–2669. doi: 10.1093/bioinformatics/bty149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jesus J.G., Giovanetti M., Rodrigues Faria N., Alcantara L.C.J. Acute vector-borne viral infection: zika and MinION surveillance. Microbiol. Spectr. 2019;7 doi: 10.1128/microbiolspec.AME-0008-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villiers E.-M. Cross-roads in the classification of papillomaviruses. Virology, Special Issue: The Papillomavirus Episteme. 2013;445:2–10. doi: 10.1016/j.virol.2013.04.023. [DOI] [PubMed] [Google Scholar]
- de Villiers E.-M., Fauquet C., Broker T.R., Bernard H.-U., zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Dutta S., Robitaille A., Olivier M., Rollison D.E., Tommasino M., Gheit T. Genome sequence of a novel human gammapapillomavirus isolated from skin. Genome Announc. 2017;5 doi: 10.1128/genomeA.00439-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S., Robitaille A., Aubin F., Fouéré S., Galicier L., Boutboul D., Luzi F., Di Bonito P., Tommasino M., Gheit T. Identification and characterization of two novel Gammapapillomavirus genomes in skin of an immunosuppressed Epidermodysplasia Verruciformis patient. Virus Res. 2018;249:66–68. doi: 10.1016/j.virusres.2018.03.003. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekström J., Bzhalava D., Svenback D., Forslund O., Dillner J. High throughput sequencing reveals diversity of Human Papillomaviruses in cutaneous lesions. Int. J. Cancer. 2011;129:2643–2650. doi: 10.1002/ijc.26204. [DOI] [PubMed] [Google Scholar]
- Gheit T. Mucosal and cutaneous human papillomavirus infections and Cancer biology. Front. Oncol. 2019;9:355. doi: 10.3389/fonc.2019.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein S., Beka L., Graf J., Klassen J.L. Evaluation of strategies for the assembly of diverse bacterial genomes using MinION long-read sequencing. BMC Genomics. 2019;20:23. doi: 10.1186/s12864-018-5381-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L., Naccache S.N., Federman S., Yu G., Mbala P., Bres V., Stryke D., Bouquet J., Somasekar S., Linnen J.M., Dodd R., Mulembakani P., Schneider B.S., Muyembe-Tamfum J.-J., Stramer S.L., Chiu C.Y. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med. 2015;7:99. doi: 10.1186/s13073-015-0220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther T., Haas L., Alawi M., Wohlsein P., Marks J., Grundhoff A., Becher P., Fischer N. Recovery of the first full-length genome sequence of a parapoxvirus directly from a clinical sample. Sci. Rep. 2017;7:3734. doi: 10.1038/s41598-017-03997-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen H.J., Liem M., Jong-Raadsen S.A., Dufour S., Weltzien F.-A., Swinkels W., Koelewijn A., Palstra A.P., Pelster B., Spaink H.P., Thillart G.Evanden, Dirks R.P., Henkel C.V. Rapid de novo assembly of the European eel genome from nanopore sequencing reads. Sci. Rep. 2017;7:7213. doi: 10.1038/s41598-017-07650-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson H., Bzhalava D., Ekström J., Hultin E., Dillner J., Forslund O. Metagenomic sequencing of “HPV-negative” condylomas detects novel putative HPV types. Virology. 2013;440:1–7. doi: 10.1016/j.virol.2013.01.023. [DOI] [PubMed] [Google Scholar]
- Karamitros T., Harrison I., Piorkowska R., Katzourakis A., Magiorkinis G., Mbisa J.L. De novo assembly of human herpes virus type 1 (HHV-1) genome, mining of non-canonical structures and detection of novel drug-resistance mutations using short- and long-read next generation sequencing technologies. PLoS One. 2016;11 doi: 10.1371/journal.pone.0157600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasianowicz J.J., Brandin E., Branton D., Deamer D.W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. U. S. A. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M.W., Rambo-Martin B.L., Wilson M.M., Ridenour C.A., Shepard S.S., Stark T.J., Neuhaus E.B., Dugan V.G., Wentworth D.E., Barnes J.R. Direct RNA sequencing of the coding complete influenza a virus genome. Sci. Rep. 2018;8:14408. doi: 10.1038/s41598-018-32615-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocjan B.J., Bzhalava D., Forslund O., Dillner J., Poljak M. Molecular methods for identification and characterization of novel papillomaviruses. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2015;21:808–816. doi: 10.1016/j.cmi.2015.05.011. [DOI] [PubMed] [Google Scholar]
- Lanfear R., Schalamun M., Kainer D., Wang W., Schwessinger B. MinIONQC: fast and simple quality control for MinION sequencing data. Bioinforma. Oxf. Engl. 2019;35:523–525. doi: 10.1093/bioinformatics/bty654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski K., Xu Y., Pullan S.T., Lumley S.F., Foster D., Sanderson N., Vaughan A., Morgan M., Bright N., Kavanagh J., Vipond R., Carroll M., Marriott A.C., Gooch K.E., Andersson M., Jeffery K., Peto T.E.A., Crook D.W., Walker A.S., Matthews P.C. Metagenomic Nanopore Sequencing of Influenza Virus Direct from Clinical Respiratory Samples. J. Clin. Microbiol. 2019;58 doi: 10.1128/JCM.00963-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Chng K.R., Boey E.J.H., Ng A.H.Q., Wilm A., Nagarajan N. INC-Seq: accurate single molecule reads using nanopore sequencing. GigaScience. 2016;5:34. doi: 10.1186/s13742-016-0140-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Giordano F., Ning Z. Oxford nanopore MinION sequencing and genome assembly. Genomics Proteomics Bioinformatics. 2016;14:265–279. doi: 10.1016/j.gpb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makałowski W., Shabardina V. Bioinformatics of nanopore sequencing. J. Hum. Genet. 2019 doi: 10.1038/s10038-019-0659-4. [DOI] [PubMed] [Google Scholar]
- McNaughton A.L., Roberts H.E., Bonsall D., de Cesare M., Mokaya J., Lumley S.F., Golubchik T., Piazza P., Martin J.B., de Lara C., Brown A., Ansari M.A., Bowden R., Barnes E., Matthews P.C. Illumina and Nanopore methods for whole genome sequencing of hepatitis B virus (HBV) Sci. Rep. 2019;9:7081. doi: 10.1038/s41598-019-43524-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medaka: Sequence correction provided by ONT Research (Accessed 27 Jun 2020). n.d.
- Moldován N., Tombácz D., Szűcs A., Csabai Z., Balázs Z., Kis E., Molnár J., Boldogkői Z. Third-generation sequencing reveals extensive polycistronism and transcriptional overlapping in a baculovirus. Sci. Rep. 2018;8:8604. doi: 10.1038/s41598-018-26955-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühr L.S.A., Eklund C., Dillner J. Towards quality and order in human papillomavirus research. Virology. 2018;519:74–76. doi: 10.1016/j.virol.2018.04.003. [DOI] [PubMed] [Google Scholar]
- Orth G. Genetics of epidermodysplasia verruciformis: insights into host defense against papillomaviruses. Semin. Immunol. 2006;18:362–374. doi: 10.1016/j.smim.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Pastrana D.V., Peretti A., Welch N.L., Borgogna C., Olivero C., Badolato R., Notarangelo L.D., Gariglio M., FitzGerald P.C., McIntosh C.E., Reeves J., Starrett G.J., Bliskovsky V., Velez D., Brownell I., Yarchoan R., Wyvill K.M., Uldrick T.S., Maldarelli F., Lisco A., Sereti I., Gonzalez C.M., Androphy E.J., McBride A.A., Van Doorslaer K., Garcia F., Dvoretzky I., Liu J.S., Han J., Murphy P.M., McDermott D.H., Buck C.B. Metagenomic discovery of 83 new human papillomavirus types in patients with immunodeficiency. mSphere. 2018;3 doi: 10.1128/mSphereDirect.00645-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. Genome sequencing. Search for pore-fection. Science. 2012;336:534–537. doi: 10.1126/science.336.6081.534. [DOI] [PubMed] [Google Scholar]
- Prazsák I., Moldován N., Balázs Z., Tombácz D., Megyeri K., Szűcs A., Csabai Z., Boldogkői Z. Long-read sequencing uncovers a complex transcriptome topology in varicella zoster virus. BMC Genomics. 2018;19:873. doi: 10.1186/s12864-018-5267-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prazsák I., Tombácz D., Szűcs A., Dénes B., Snyder M., Boldogkői Z. Full genome sequence of the western reserve strain of vaccinia virus determined by third-generation sequencing. Genome Announc. 2018;6 doi: 10.1128/genomeA.01570-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rector A., Tachezy R., Van Ranst M. A sequence-independent strategy for detection and cloning of circular DNA virus genomes by using multiply primed rolling-circle amplification. J. Virol. 2004;78:4993–4998. doi: 10.1128/jvi.78.10.4993-4998.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner J.E., Balijepalli A., Robertson J.W.F., Campbell J., Suehle J., Kasianowicz J.J. Disease detection and management via single nanopore-based sensors. Chem. Rev. 2012;112:6431–6451. doi: 10.1021/cr300381m. [DOI] [PubMed] [Google Scholar]
- Sasani T.A., Cone K.R., Quinlan A.R., Elde N.C. Long read sequencing reveals poxvirus evolution through rapid homogenization of gene arrays. eLife. 2018;7 doi: 10.7554/eLife.35453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schowalter R.M., Pastrana D.V., Pumphrey K.A., Moyer A.L., Buck C.B. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe. 2010;7:509–515. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombácz D., Balázs Z., Csabai Z., Snyder M., Boldogkői Z. Long-read sequencing revealed an extensive transcript complexity in Herpesviruses. Front. Genet. 2018;9:259. doi: 10.3389/fgene.2018.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombácz D., Prazsák I., Szucs A., Dénes B., Snyder M., Boldogkoi Z. Dynamic transcriptome profiling dataset of vaccinia virus obtained from long-read sequencing techniques. GigaScience. 2018;7 doi: 10.1093/gigascience/giy139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombácz D., Moldován N., Balázs Z., Gulyás G., Csabai Z., Boldogkői M., Snyder M., Boldogkői Z. Multiple long-read sequencing survey of herpes simplex virus dynamic transcriptome. Front. Genet. 2019;10:834. doi: 10.3389/fgene.2019.00834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasino M. The human papillomavirus family and its role in carcinogenesis. Semin. Cancer Biol. 2014;26:13–21. doi: 10.1016/j.semcancer.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Tommasino M. The biology of beta human papillomaviruses. Virus Res. 2017;231:128–138. doi: 10.1016/j.virusres.2016.11.013. [DOI] [PubMed] [Google Scholar]
- Tyler A.D., Mataseje L., Urfano C.J., Schmidt L., Antonation K.S., Mulvey M.R., Corbett C.R. Evaluation of oxford nanopore’s MinION sequencing device for microbial whole genome sequencing applications. Sci. Rep. 2018;8:10931. doi: 10.1038/s41598-018-29334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida Y., Kouyama J.-I., Naiki K., Uemura H., Tsuji S., Sugawara K., Nakao M., Motoya D., Nakayama N., Imai Y., Tomiya T., Mochida S. A case of genotype-3b hepatitis C virus in which the whole genome was successfully analyzed using third-generation nanopore sequencing. Hepatol. Res. Off. J. Jpn. Soc. Hepatol. 2019;49:1083–1087. doi: 10.1111/hepr.13339. [DOI] [PubMed] [Google Scholar]
- Vanmechelen B., Bertelsen M.F., Rector A., Van den Oord J.J., Laenen L., Vergote V., Maes P. Identification of a novel species of papillomavirus in giraffe lesions using nanopore sequencing. Vet. Microbiol. 2017;201:26–31. doi: 10.1016/j.vetmic.2016.12.035. [DOI] [PubMed] [Google Scholar]
- Vanmechelen B., Rector A., Maes P. Virus hunting: discovery of new episomal circular viruses by rolling circle techniques. Curr. Protoc. Microbiol. 2017;44 doi: 10.1002/cpmc.23. 1E.12.1-1E.12.18. [DOI] [PubMed] [Google Scholar]
- Wawina-Bokalanga T., Vanmechelen B., Martí-Carreras J., Vergote V., Vermeire K., Muyembe-Tamfum J.-J., Ahuka-Mundeke S., Maes P. Complete genome sequence of a new ebola virus strain isolated during the 2017 likati outbreak in the Democratic Republic of the Congo. Microbiol. Resour. Announc. 2019;8 doi: 10.1128/MRA.00360-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R.R., Judd L.M., Holt K.E. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 2019;20:129. doi: 10.1186/s13059-019-1727-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Lewandowski K., Lumley S., Pullan S., Vipond R., Carroll M., Foster D., Matthews P.C., Peto T., Crook D. Detection of viral pathogens with multiplex nanopore MinION sequencing: Be careful with cross-talk. Front. Microbiol. 2018;19:2225. doi: 10.3389/fmicb.2018.02225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yinda C.K., Seifert S.N., Macmenamin P., van Doremalen N., Kim L., Bushmaker T., de Wit E., Quinones M., Munster V.J. A novel field-deployable method for sequencing and analyses of henipavirus genomes from complex samples on the MinION platform. J. Infect. Dis. 2019 doi: 10.1093/infdis/jiz576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data are available in the Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra), under BioProject PRJNA673786. All the code can be retrieved on the IARC bioinformatics platform on GitHub (https://github.com/IARCbioinfo).