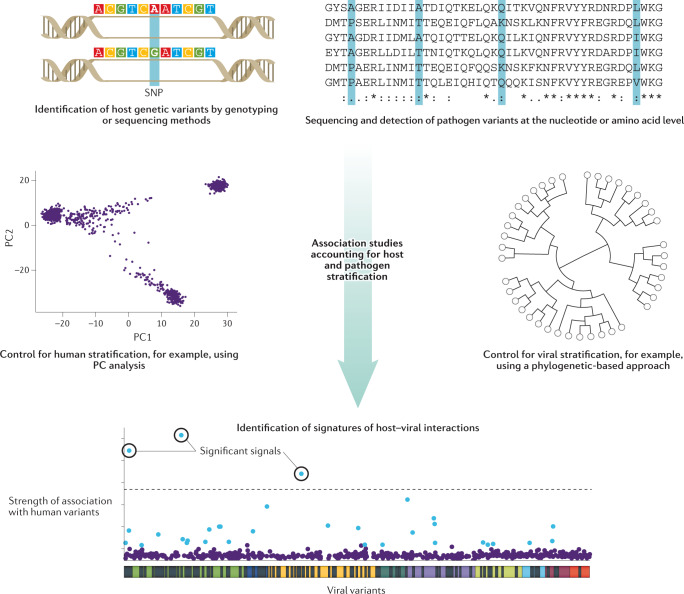
Fig. 4. Detecting genomic signatures of host–pathogen interactions in matched host and virus samples.
First, genetic variants in the host (human) and pathogen (viral) genomes are identified from genome-wide genotyping or sequencing data and catalogued. A genome-wide search for associations between human polymorphisms and viral variants is then performed, which needs to consider the risk of systematic signal inflation owing to population stratification. On the human side, this can be addressed by methods that infer genetic ancestry, such as principal component (PC) analysis followed by the inclusion of the top PCs as covariates, or by the use of mixed models that incorporate the full covariance structure of the study population130. On the pathogen side, various phylogenetic-based and model-based approaches have been proposed85. Significant associations, after correction for multiple testing, reveal the loci involved in host–pathogen genomic conflicts. The plot showing the signatures of viral–host interactions is adapted from ref.84, CC BY 3.0 (https://creativecommons.org/licenses/by/3.0/).