Fig. 6.
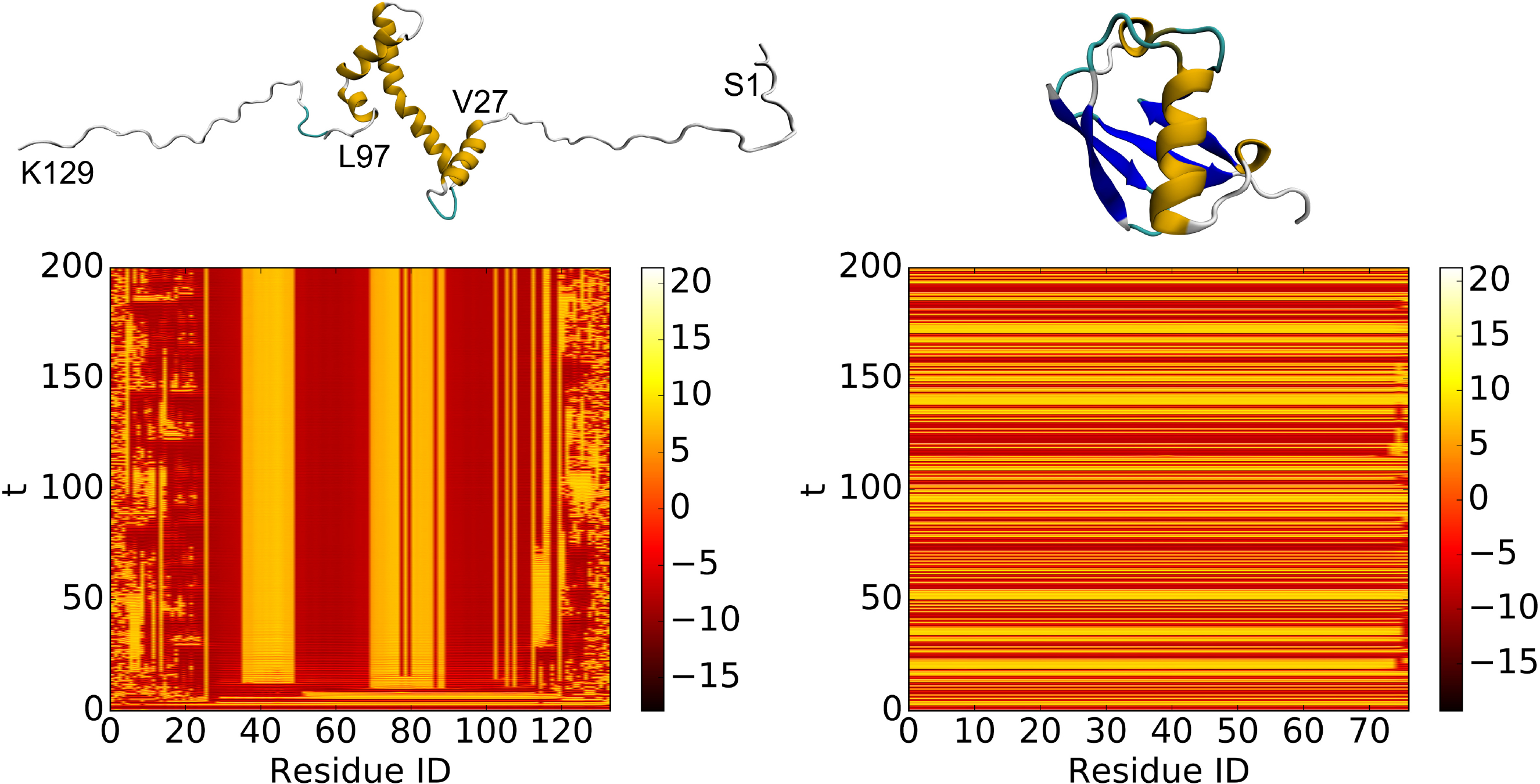
Left: partially disordered protein, model 1 of PDB:2RVQ. Right: well folded protien, PDB:1UBQ. The ui,1 value of each dynamical system is plotted as heatmap. The Lorenz system defined in Eq. (2) is used with the parameters δ = 10, γ = 28, β = 8/3. The coupling matrix A defined in Eq. (11) has parameters μ = 14, κ = 2. The coupled system defined in Eq. (1) has parameters Γ = I3 and ϵ = 12. The system is initialized with a random value between 0 and 1 and is simulated from t = 0 to t = 200 with step size h = 0.01. The system is numerically solved using the 4-th order Runge-Kutta method. It can be seen from the heatmaps that the oscillators corresponding to the disordered regions behave asynchronously.
