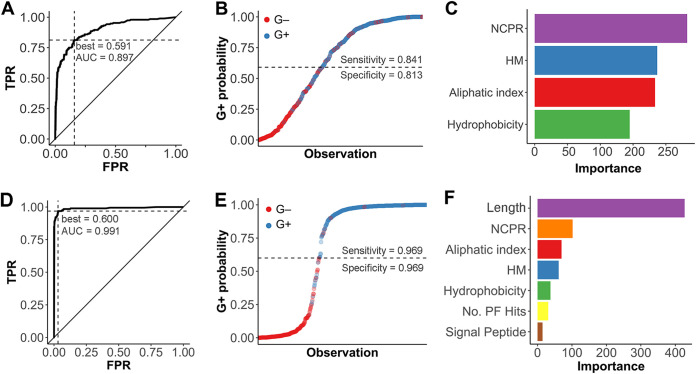
FIG 9.
Random forest prediction and classification of Gram groups of bacterial hosts based on physicochemical properties of lysins (A, B, and C) or on those properties plus others relative to lysin architecture (D, E, and F). (A and D) ROC curves of the random forest predictive models (TPR, true-positive rate; FPR, false-positive rate). ROC best points of positive-group (G+) probability for outcome maximization are presented, as well as the AUCs. (B and E) Random forest castings of bacterial host Gram group on the testing subset of lysin sequences. The dashed lines represent the G+ probability threshold for classification based on the respective ROC best points. (C and F) Importance (i.e., mean Gini index decrease for each variable) of each of the four descriptors used for classification within each model. HM, hydrophobic moment.