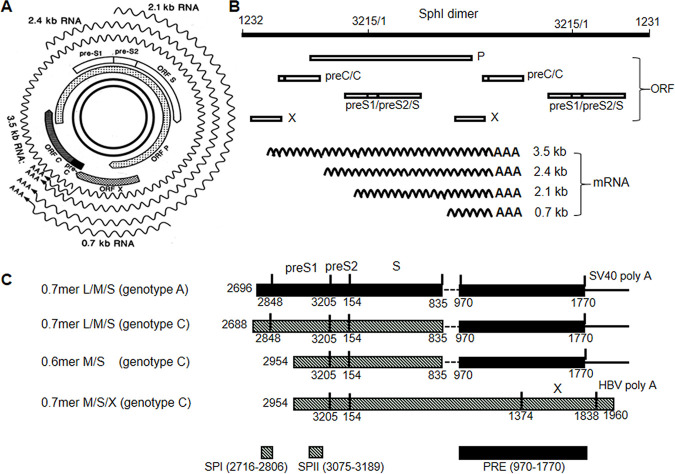
FIG 1.
Schematic view of HBV RNA transcription, envelope protein expression, and genome replication from cccDNA, SphI dimer, and subgenomic expression constructs. (A) From the inside out are the double-stranded HBV genome as cccDNA, four overlapping genes or open reading frames (ORFs), and four sizes of coterminal RNAs (wavy lines). Please note that the 5′ end of the 3.5-kb RNA overlaps the 3′ end of all subgenomic RNAs. (B) The SphI dimer of genotype C as a functional equivalent of cccDNA. The top line is two tandem copies of the 3,215-nt HBV genome joined by the unique SphI site (not shown). Below are the four ORFs, and at the bottom are four sizes of HBV RNAs (wavy lines). Here, the transcription unit for 2.1-kb RNA lies downstream of those for 3.5- and 2.4-kb RNAs but upstream of that for 0.7-kb RNA. (C) Subgenomic expression constructs for HBV envelope proteins. The 0.7-mer L/M/S construct and the 0.6-mer M/S construct of geno27.2 of genotype C were derived from the 0.7-mer L/M/S construct of genotype A by replacement of the 5′ end of the HBV sequence. There is a gap of 136 nt in the HBV sequence, and the X gene (nucleotides 1771 to 1838) is incomplete at the 3′ end. Also, the poly(A) signal is derived from SV40. The 0.7-mer M/S/X construct of geno27.2 was generated by inserting a continuous HBV DNA fragment into the pBluescript SK(−) vector. It has an intact X gene and the poly(A) signal from HBV (nucleotides 1916 to 1921). The locations of the SPI and SPII promoters and the posttranscriptional regulatory element (PRE) are indicated.