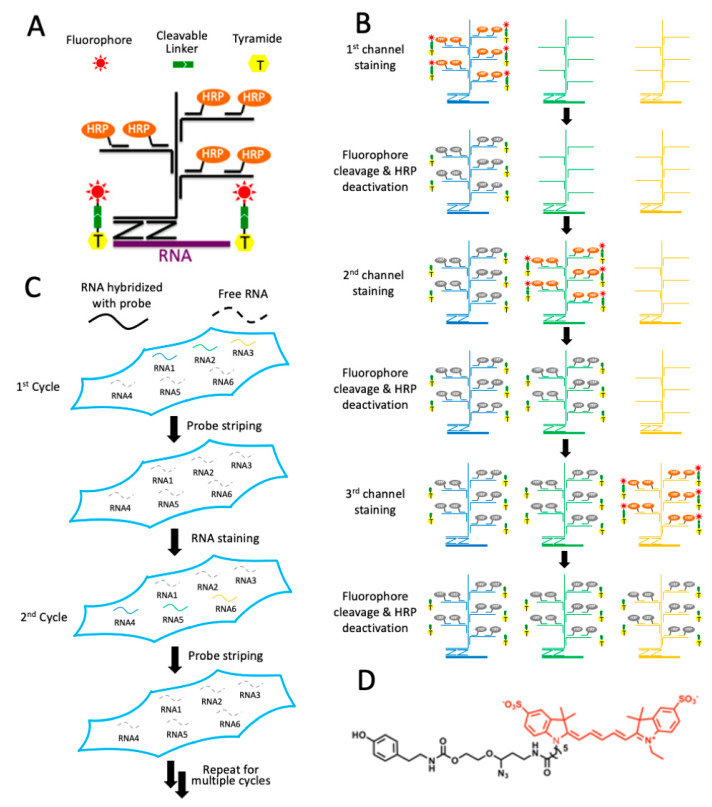
Figure 1.
Highly sensitive and multiplexed in situ RNA profiling with cleavable fluorescent tyramide (CFT). (A) In each cycle, multiple transcripts of interest are hybridized by oligonucleotide probes in pairs, which recruit oligonucleotide amplification probes. With further signal amplification by horseradish peroxidase (HRP), the target RNA is stained with CFT. (B) After imaging, the fluorophores are chemically cleaved and HRP is deactivated, allowing other RNA targets to be stained in the same cycle. (C) After all the RNA targets are stained in a cycle, all the hybridized probes are stripped. Through reiterative cycles of RNA staining and probe stripping, a large number of transcripts can be profiled in single cells in situ. (D) Structure of CFT, tyramide-N3-Cy5.