ABSTRACT
Vigilin (Vgl1) is essential for heterochromatin formation, chromosome segregation, and mRNA stability and is associated with autism spectrum disorders and cancer: vigilin, for example, can suppress proto-oncogene c-fms expression in breast cancer. Conserved from yeast to humans, vigilin is an RNA-binding protein with 14 tandemly arranged nonidentical hnRNP K-type homology (KH) domains. Here, we report that vigilin depletion increased cell sensitivity to cisplatin- or ionizing radiation (IR)-induced cell death and genomic instability due to defective DNA repair. Vigilin depletion delayed dephosphorylation of IR-induced γ-H2AX and elevated levels of residual 53BP1 and RIF1 foci, while reducing Rad51 and BRCA1 focus formation, DNA end resection, and double-strand break (DSB) repair. We show that vigilin interacts with the DNA damage response (DDR) proteins RAD51 and BRCA1, and vigilin depletion impairs their recruitment to DSB sites. Transient hydroxyurea (HU)-induced replicative stress in vigilin-depleted cells increased replication fork stalling and blocked restart of DNA synthesis. Furthermore, histone acetylation promoted vigilin recruitment to DSBs preferentially in the transcriptionally active genome. These findings uncover a novel vigilin role in DNA damage repair with implications for autism and cancer-related disorders.
KEYWORDS: vigilin, DNA repair, Rad51, homologous recombination, histone acetylation, replicative stress, autism-related disorders, cancer
INTRODUCTION
The vigilin-coding gene, a high-density lipoprotein binding protein gene (HDLBP), has been identified as one of >350 genes associated with cancer and autistic spectrum disorder (ASD) susceptibility (1–6). In mouse models, predisposition to ASD has been linked to haploinsufficiency in ASD susceptibility genes such as Nuak (7), Nbea (8), and Sh3rf2 (9), and it has been postulated that HDLBP mutant proband haploinsufficiency also predisposes to ASD (1). Functional clustering of the >350 mutated autism target genes reveals strong enrichment for genes related to fragile X mental retardation protein (FMRP) and β-catenin/chromatin remodeling networks (2, 5). As a multi-KH domain-containing protein, FMRP shares structural similarities with vigilin, and its loss-of-function mutations lead to fragile X syndrome, a mental retardation condition that, like ADS, displays deregulation of synaptic pathways (10). As with FMRP, vigilin is involved in mRNA transport to the cytoplasm (11), suggesting that the association between HDLBP haploinsufficiency and ASD may stem in part from altered mRNA transport.
Vigilin is a ubiquitous cytoplasmic and nuclear RNA-binding protein (RBP) that is evolutionarily conserved in species ranging from the yeast Saccharomyces cerevisiae (Scp160) to Drosophila (DDP1) and vertebrates (vigilin) (12, 13). It belongs to a large family of multiple KH domain proteins, collectively known as vigilins, where the KH domains can mediate protein-protein and protein-nucleic acid interactions important for RNA transport and metabolism (14–19). Vigilin interacts with the CCCTC-binding factor (CTCF) and acts in regulating the imprinted genes Igf2 and H19 (20). Vigilin has also been shown to be associated with autism and brachymetaphalangy (21, 22), whereas a 2q37 deletion syndrome that results in downregulation of vigilin causes an Albright hereditary osteodystrophy-like phenotype, characterized by developmental delay, mental retardation, and brachymetaphalangy (1).
In Drosophila, the vigilin homologue DDP1 localizes to heterochromatin, and its depletion alters nuclear morphology and causes defects in pericentromeric heterochromatin formation, with reduced H3K9 methylation and decreased heterochromatin protein 1 (HP1) recruitment. Furthermore, DDP1 depletion also suppresses heterochromatin-induced position effect variegation and induces HP1 mislocalization (23, 24). In S. cerevisiae, deletion of the vigilin homologue Scp160p causes chromosome segregation defects, which could be rescued by overexpressing Drosophila DDP1 (25). The vigilin homologue Vgl1 in Schizosaccharomyces pombe is essential for heterochromatin-mediated gene silencing, and its deletion leads to loss of H3K9 methylation and subsequent silencing defects at centromeres and telomeres (26).
In addition to their roles in chromatin condensation and gene silencing, heterochromatin-associated proteins play important roles in DNA repair (27). For example, HP1, a constitutive component of heterochromatin, is involved in the DNA damage response (28). Heterochromatin protein 1 isoforms are recruited to UV-induced DNA lesions, oxidative lesions, and DNA breaks (29). Loss of HP1 renders nematodes highly sensitive to DNA damage, and mice lacking HP1 show cellular genomic instability (29–31).
Vigilin suppresses c-fms mRNA expression by downregulating mRNA stability and inhibiting its translation (6). Although, vigilin was first described decades ago, its role in cancer and underlying mechanisms of action are still largely unknown. Given the importance of vigilin for heterochromatin organization, we examined Vgl1-deficient S. pombe cells for sensitivity to DNA-damaging agents and found the cells were hypersensitive to the DNA-damaging agent bleomycin. Since genomic instability is the hallmark of cancer (32, 33), we expanded our initial result in yeast to determine the impact of vigilin on DNA repair in human cells. Here, we report that depletion of vigilin affects DNA double-strand break (DSB) repair as well as stalled fork repair in human cell lines and thus uncover a novel role for vigilin in maintaining genomic stability.
RESULTS
Vigilin depletion decreases cell survival and increases genomic instability.
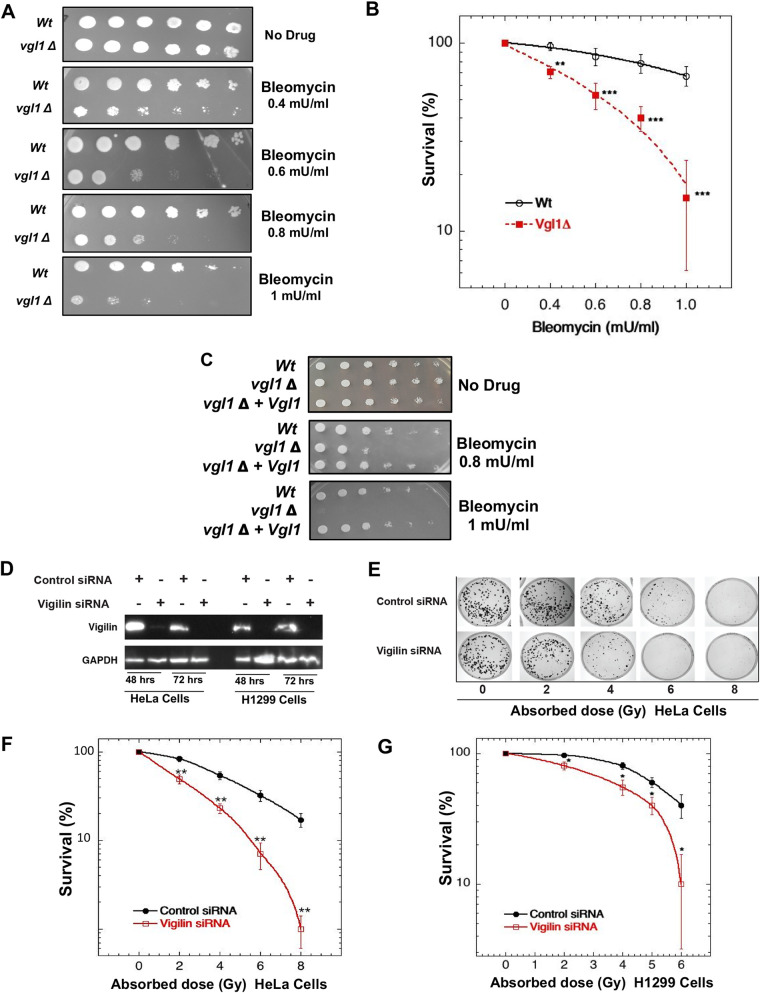
Heterochromatin integrity is important for genome stability, and heterochromatin-associated proteins can also play a direct role in DNA damage repair (28, 29, 31, 34). We tested whether Vgl1 contributes to the DNA damage response by treating wild-type and Vgl1-deleted S. pombe cells with the DNA double-strand break-inducing agent bleomycin. Cells lacking Vgl1 were hypersensitive to bleomycin and displayed reduced cell survival compared to wild-type cells (Fig. 1A and B). Ectopic expression of Vgl1 in Vgl1-deficient cells restored resistance to bleomycin (Fig. 1C), confirming that Vgl1 plays a role in cell survival post-bleomycin treatment.
FIG 1.
Loss of vigilin decreases cell survival postirradiation. (A) Tenfold serial dilution plating assays of wild-type (Wt) and vgl1Δ S. pombe cells showing bleomycin sensitivity. (B) Cell survival assay of wild-type and vgl1Δ S. pombe cells grown in the presence of bleomycin. (C) Ectopic expression of Vgl1 in cells with Vgl1 deleted rescues bleomycin sensitivity. (D) Western blot showing the knockdown of vigilin in HeLa and H1299 cell lines using vigilin-specific and control siRNAs. (E to G) Clonogenic cell survival assays after irradiation with different doses of IR in HeLa and H1299 cells with and without knockdown of vigilin. The error bars represent the standard deviations from three independent experiments.
To test whether vigilin, the human homologue of Vgl1, is similarly protective against DNA-damaging agents, we depleted vigilin in H1299 and HeLa cells and then measured clonogenic cell survival after ionizing radiation (IR) exposure. Vigilin depletion significantly reduced cell survival postirradiation in both H1299 and HeLa cells (Fig. 1D to G). Identical survival results were obtained using different vigilin small interfering RNAs (siRNAs) or short hairpin RNAs (shRNAs), confirming the original phenotype (data not shown). Overall, the results indicate that vigilin plays a role in DNA damage response.
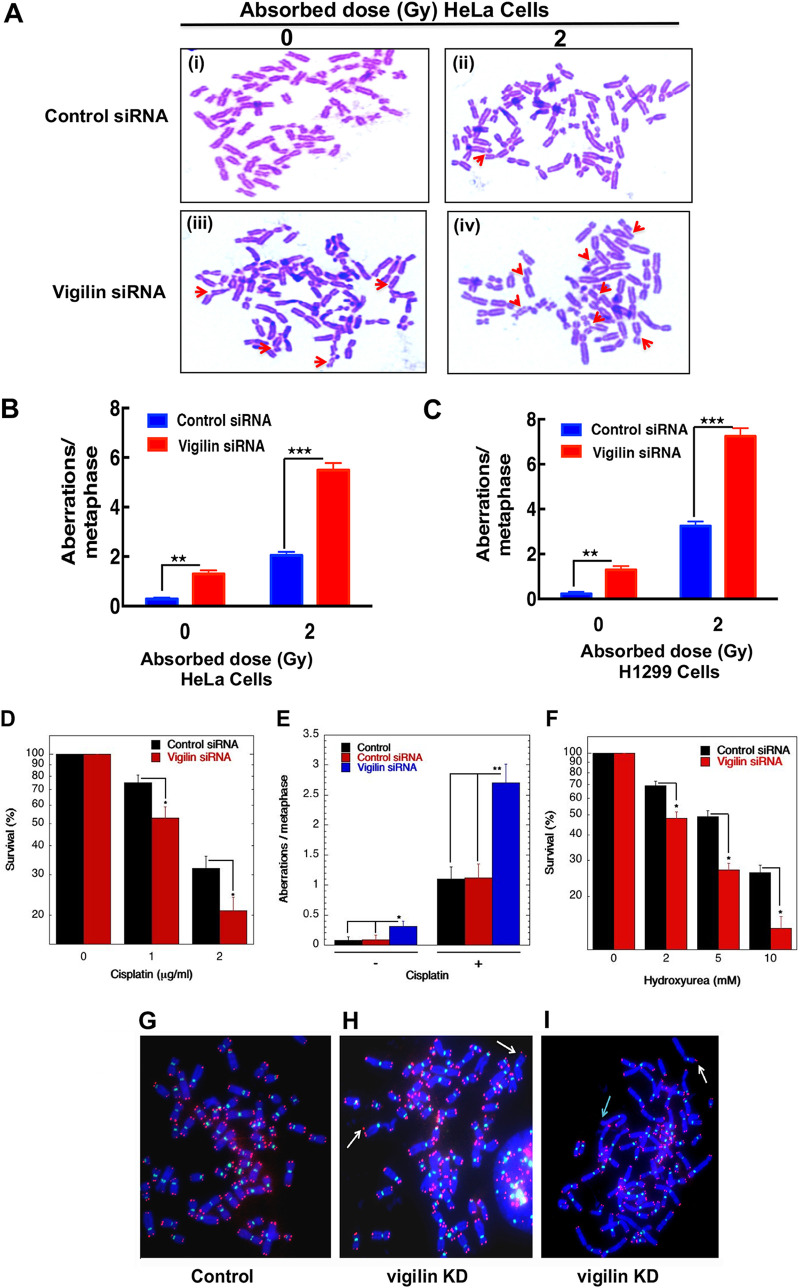
To determine whether the observed IR sensitivity was due to impaired DNA damage repair, we measured chromosomal aberrations in H1299 and HeLa cells with and without vigilin depletion. Unirradiated vigilin knockdown cells had increased endogenous levels of various types of chromosomal aberrations, including breaks, fragments, radials, etc., in comparison to control cells with vigilin, indicative of spontaneous genomic instability (Fig. 2A to C). Furthermore, the levels of IR-induced chromosomal aberrations were also higher in vigilin-depleted cells than in irradiated cells with vigilin (Fig. 2A to C). These results suggest that vigilin is essential for DNA damage repair as its absence increased spontaneous defects, as well as enhanced IR-induced chromosomal aberrations. To establish whether vigilin also has a role in repair of DNA cross-links, we treated control and vigilin-depleted cells with cisplatin or hydroxyurea (HU) and measured cell survival and chromosomal aberrations at metaphase. Cells lacking vigilin displayed reduced cell survival upon cisplatin and hydroxyurea treatment and had significantly increased chromosomal aberrations compared to control cells (Fig. 2D to F). These results suggest that vigilin plays a role in the repair of DNA damage.
FIG 2.
Chromosome aberration analysis. (A) Representative images of metaphase spread of cells in control and vigilin knockdown HeLa cells showing increased spontaneous and IR-induced metaphase chromosome aberrations. (B and C) Quantification of chromosome aberrations seen in HeLa and H1299 cells, respectively, with and without depletion of vigilin. Quantification represents aberrations from 30 metaphase spreads of three independent experiments. (D to F) Clonogenic cell survival assay and metaphase chromosome aberrations of vigilin-depleted HeLa cells treated with the indicated concentrations of cisplatin and hydroxyurea. Vigilin-depleted cells show decreased survival and increased chromosome aberrations upon treatment with cisplatin and hydroxyurea compared to control cells. The error bars represent the standard deviations from three independent experiments. (G) Human metaphase chromosomes showing telomeres in red and centromeres in green detected by FISH using specific probes in control and vigilin knockdown cells. (H and I) Metaphase chromosomes from vigilin-depleted cells showing dicentrics indicated by white arrows and loss of telomere signals indicated by green arrow.
Genomic stability is dependent partly on telomere integrity, such that telomere dysregulation can lead to telomere fusions (30, 35–39). To assess whether telomere stability was compromised by vigilin depletion, we performed fluorescent in situ hybridization (FISH) on metaphase chromosome spreads using a telomere-specific Cy3-labeled (CCCTAA)3 peptide nucleic acid probe. In vigilin-depleted cells, there was loss of telomeric signal, along with aneuploidy and dicentric or multicentric chromosome formation (Fig. 2G to I), which could be due to breaks or shortened telomeres. These results confirm that vigilin plays a crucial role in maintaining genome stability.
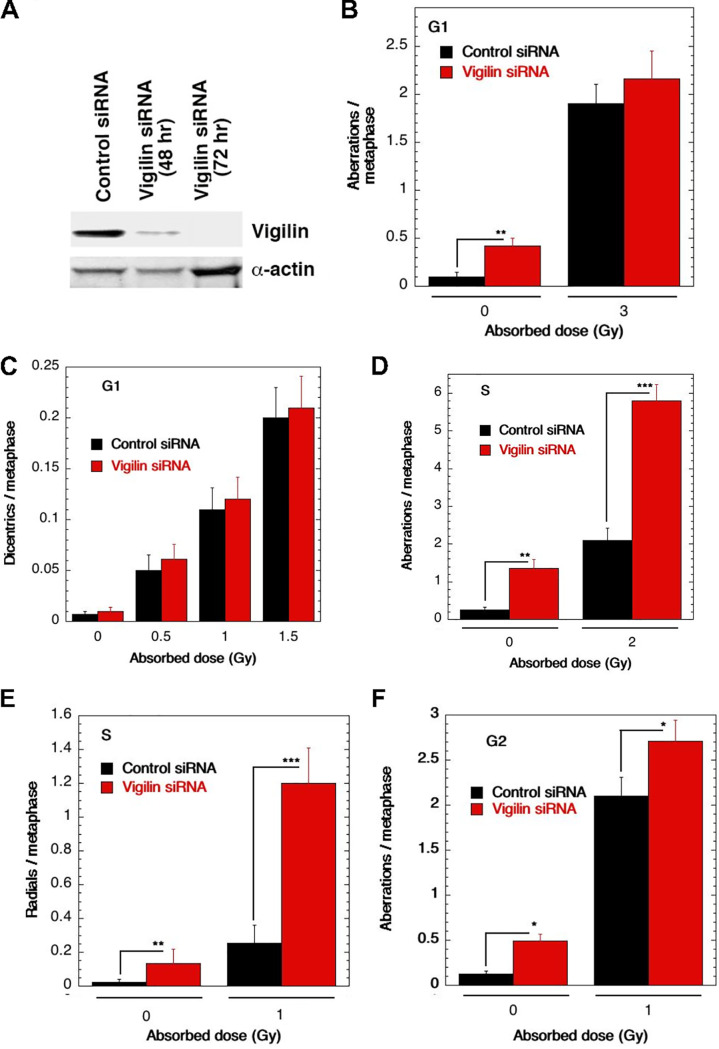
To delineate the cell cycle-specific chromosomal aberrations, we measured the frequency of chromosomal and chromatid-type aberrations observed at metaphase (40). G1-specific chromosome aberrations detected at metaphase are mostly of the chromosomal type with dicentrics (41). S-phase-type aberrations ascertained at metaphase are chromosomal; in addition, chromatid type and G2-type aberrations observed at metaphase are mostly the chromatid type. To determine G1-type chromosome damage, cells were exposed to 3 Gy of IR, and aberrations were scored at metaphase as previously described (41, 42). No difference in residual IR-induced G1 chromosomal aberrations was seen at metaphase with or without vigilin knockdown (Fig. 3A to C). To determine S-phase-specific aberrations, we first determined the time needed for S-phase cells to reach metaphase after IR treatment. Exponentially growing cells were labeled with bromodeoxyuridine (BrdU) for 30 min as previously described (30) and then irradiated with 2 Gy. A previously described procedure was used to determine when S-phase cells reached metaphase (30). Cells were treated with 2 Gy of IR, and metaphases were collected 4 to 5 h postirradiation. Cells with vigilin knockdown had a higher frequency of chromosomal and chromatid aberrations, specifically radials (Fig. 2D and E). G2-type aberrations were scored in cells treated with 1 Gy followed by metaphase collection 1 h later and found to be higher in vigilin knockdown cells (Fig. 3F). These observations suggest a role for vigilin in repairing chromosome damage in the S phase as well as in the G2 phase of the cell cycle.
FIG 3.
Analysis of cell cycle-specific chromosome damage in HCT116 cells with or without depletion of vigilin. (A) Western blot showing depletion of vigilin after 48 and 72 h of control and vigilin-specific siRNA tranfection. (B) Cells in the plateau phase were irradiated with 3 Gy, incubated for 24 h postirradiation, and then subcultured, and metaphases were analyzed. G1-type aberrations were examined at metaphase. All categories of asymmetric chromosome aberrations induced by IR were scored: dicentrics, centric rings, interstitial deletions/acentric rings, and terminal deletions. The frequencies of chromosomal aberrations were identical for cells with or without depletion of vigilin, indicating normal repair of G1-phase-specific chromosome damage. (C) Dicentrics, which formed mostly in G1, were analyzed after 3 Gy of IR exposure, and no differences were found in cells with and without depletion of vigilin. (D) Cells in the exponential phase were irradiated with 2 Gy. Metaphases were harvested at 4 h after irradiation, and S-phase types of chromosomal aberrations were scored. Cells with vigilin knockdown showed significant differences in chromosomal aberration frequencies compared to control cells. (E) Radials were analyzed after IR exposure. (F) Cells in the exponential phase were irradiated with 1 Gy. Metaphases were harvested following irradiation, and G2-type chromosomal aberrations were monitored. Cells with vigilin knockdown showed modest increase in frequencies of chromosomal aberrations compared to control cells. Each experiment was repeated three times, and for each experiment, 300 metaphases were scored.
Absence of vigilin alters the DNA damage response.
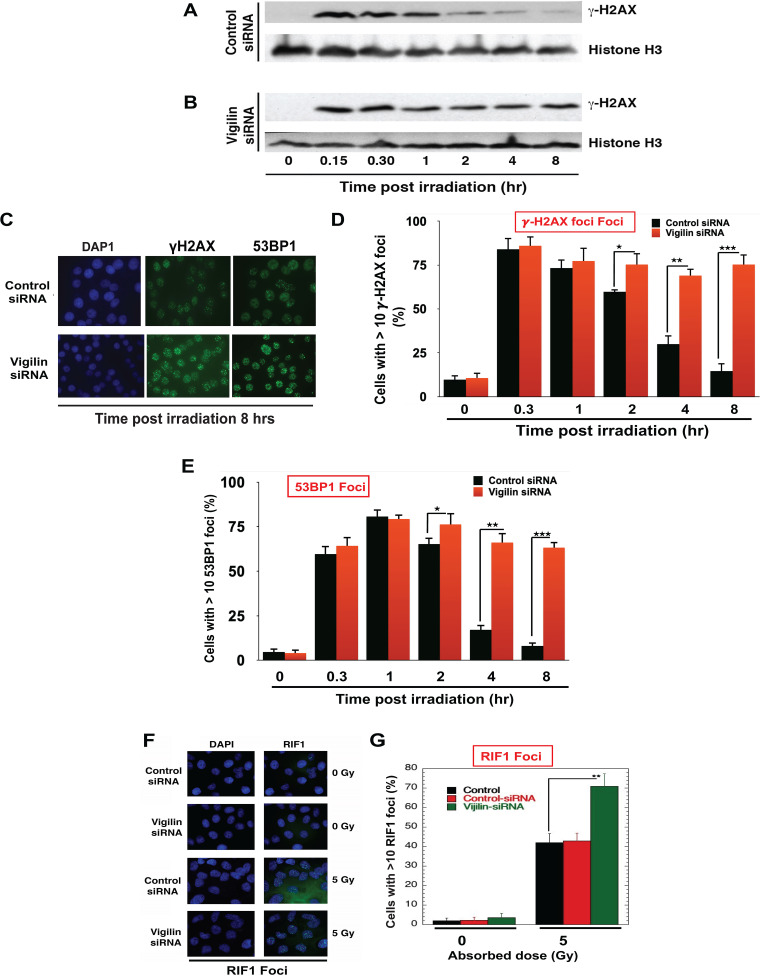
One of the earliest events in the DNA damage response is the phosphorylation of histone H2AX on serine 139 (γ-H2AX) by ATM/ATR kinases. This phosphorylation mark acts as a binding site for subsequent repair proteins, including 53BP1, and is also important for DNA repair pathway activation (43–49). We therefore tested whether vigilin depletion impacted H2AX phosphorylation levels. At 15 min postirradiation, γ-H2AX levels in vigilin-depleted cells were similar to that in control cells, as measured by Western blotting (Fig. 4A and B). However, by 2 h postirradiation, γ-H2AX levels in control cells had significantly declined, indicating completion of DNA damage repair, while γ-H2AX levels in vigilin-depleted cells did not decrease, even after 8 h, suggesting a defect in processing DNA damage (Fig. 4A and B). In agreement with the Western data, IR-induced γ-H2AX foci did not decline even by 8 h in vigilin-depleted cells, further indicating defective DNA repair (Fig. 4C and D).
FIG 4.
Depletion of vigilin alters the DNA damage response. (A and B) HeLa cells were transfected with either control siRNA or vigilin siRNA followed by irradiation with 2 Gy and then analyzed by Western blotting for the appearance/disappearance of γ-H2AX after release at different time points. H3 was used as a loading control. (C) Control and vigilin-depleted cells were irradiated with 4 Gy, fixed after IR treatment, and stained for γ-H2AX and 53BP1 foci. (D and E) Quantification of percentage of cells with γ-H2AX and 53BP1 foci in control and vigilin knockdown cells at different time points postirradiation. For each time point, 100 cells were analyzed, and the number of cells with more than 10 foci was plotted against time. (F and G) HeLa cells were irradiated with 5 Gy, and RIF1 foci were quantified for RIF1 foci in control and vigilin-depleted HeLa cells with and without IR treatment. Vigilin-depleted cells show higher levels of residual γ-H2AX, 53BP1, and RIF1 foci postirradiation compared to control cells. Error bars represent standard deviations from three independent experiments.
Similar to the results for γ-H2AX foci, the initial appearance of 53BP1 foci in vigilin-depleted cells was equal to that in control cells. However, at 8 h postirradiation 53BP1 foci in vigilin-depleted cells was higher than in control cells, indicating defective recruitment of downstream repair factors (Fig. 4C to E). Since we observed higher levels of residual 53BP1 in vigilin-depleted cells, we also measured RIF1, the first effector protein of 53BP1 foci, at DSBs and found increased RIF1 foci postirradiation (5 Gy) in vigilin-depleted cells (Fig. 4F and G). Higher levels of residual γ-H2AX, 53BP1, and RIF1 foci all suggest that DNA damage repair is impaired or compromised in vigilin-deficient cells.
Vigilin depletion affects DNA DSB repair by homologous recombination.
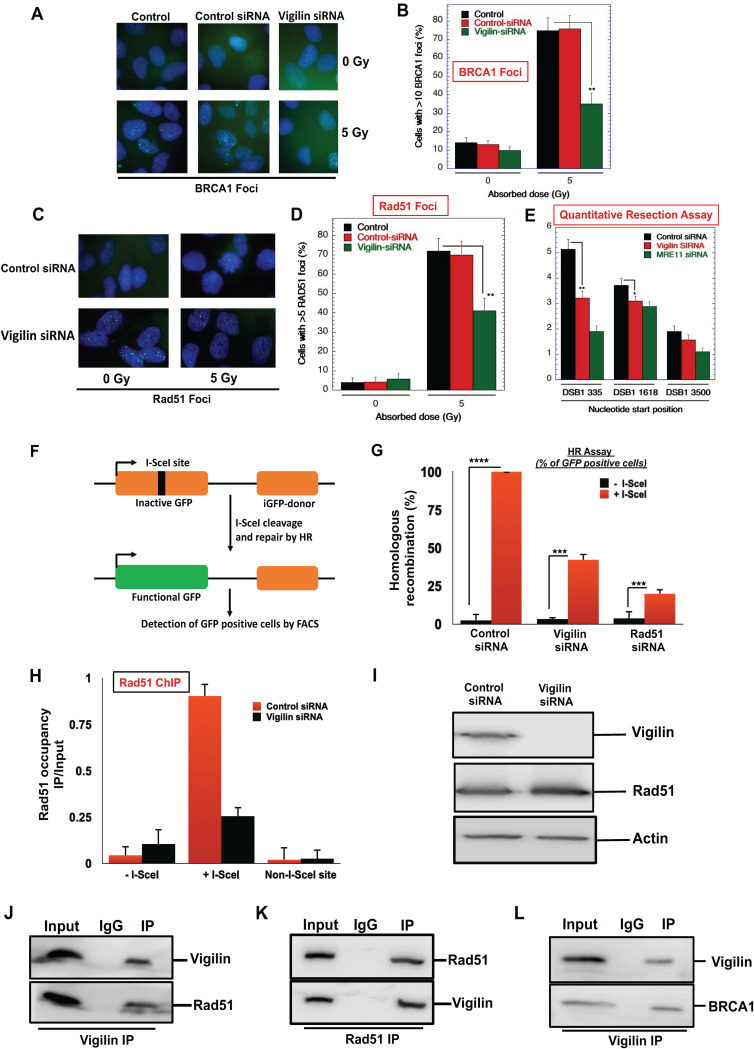
Double-strand break repair by homologous recombination (HR) involves 53BP1 replacement by BRCA1 to facilitate MRN (MRE11-RAD50-NBS1)-mediated DNA end resection (50–52). This is followed by replication protein A (RPA) loading onto the resulting single-stranded DNA (ssDNA) and then displacement by and polymerization of RAD51 to initiate the homologous pairing and strand exchange steps (51). Since vigilin depletion increased the number of cells with residual 53BP1 and RIF1 foci, we tested whether depletion also impacted BRCA1 focus formation. After irradiation, vigilin-depleted cells formed fewer BRCA1 foci compared to cells with vigilin (Fig. 5A and B). BRCA1 facilitates RAD51 recruitment, and vigilin depletion was observed to decrease formation of RAD51 foci postirradiation (5 Gy) (Fig. 5C and D). We then tested whether the decreased formation of BRCA1 and RAD51 foci in vigilin-depleted cells was the result of decreased ssDNA formation by resection. In a quantitative resection assay using the estrogen receptor (ER)-AsiSI system and MRE11 depletion as a positive control (53–55), decreased ssDNA formation was observed in vigilin-depleted cells compared to control cells (Fig. 5E).
FIG 5.
Loss of vigilin impairs the homologous recombination repair pathway. (A to D) Quantification of cells for IR-induced BRCA1 and RAD51 focus formation in control and vigilin-depleted cells. (E) Quantitative resection assay showing the percentage of ssDNA formation in control and vigilin-depleted cells. The assay was done using the ER-AsiSI system, as described in Materials and Methods. Vigilin-depleted cells show significant reduction in ssDNA formation, Mre11-depleted ER-AsiSI U2OS cells were used as a positive control. Error bars are from three independently performed experiments. (F) Schematic diagram of the HR DR-GFP reporter assay cassette used to analyze homologous recombination. Homologous recombination was assessed using a direct repeat-green fluorescent protein (DR-GFP) cassette stably integrated at a single locus in H1299 cells. The DR-GFP gene is inactivated by insertion of an I-SecI site containing a stop codon. The downstream iGFP gene has 0.8-kb sequence homology to direct the repair of an I-SecI-cleaved inactive GFP, which results in the restoration of a functional GFP. (G) Cells were transfected with the indicated siRNAs for 36 h, followed by transfection with I-SecI plasmid to induce DSB. Cells were assessed after 48 h by fluorescence-activated cell sorter (FACS) analysis for GFP expression. Results indicate percentage of GFP-positive cells normalized to control cells from three independent experiments. (H) Vigilin depletion impairs recruitment of Rad51 to DSB sites. Cells were depleted with either control siRNA or vigilin siRNA, followed by induction of DSB by I-SceI. Rad51 occupancy was analyzed by chromatin immunoprecipitation at the I-SceI site in cells with and without the depletion of vigilin. The results represent standard errors from three independent experiments. (I) Rad51 protein levels in control and vigilin-depleted cells indicating that impairment of Rad51 recruitment to DSBs in cells lacking vigilin is not due to change in Rad51 protein levels. (J) Coimmunoprecipitation experiments showing vigilin interaction with Rad51. Vigilin was immunoprecipitated using vigilin-specific antibody; immunoprecipitated fractions were analyzed by Western blotting using anti-Rad51 antibodies. (K) Rad51 antibody was used for immunoprecipitation, and immunoprecipitated fractions were analyzed by Western blotting using antivigilin antibodies. (L) Immunoprecipitation experiment showing vigilin interaction with BRCA1. Vigilin was immunoprecipitated using a vigilin-specific antibody; immunoprecipitated fractions were analyzed by Western blotting using anti-BRCA1 antibodies.
The cumulative data of higher residual γ-H2AX, 53BP1, and RIF1 foci and lower RAD51 and BRCA1 foci in vigilin-depleted cells indicate defective repair of DNA DSBs by the homologous recombination (HR) pathway. To further test this conclusion, we used an engineered cell line that expresses green fluorescent protein (GFP) only upon reconstitutive repair of I-SceI-induced DSBs by HR (Fig. 5F). An I-SceI expression vector was transfected into MCF7 cells containing a stably integrated pDR-GFP plasmid (MCF7 direct repeat [DR]-GFP cells), with and without vigilin depletion, and flow cytometry was used to quantify GFP-positive cells. After I-SceI transfection, 50% fewer GFP-positive cells were detected after vigilin depletion compared to control cell levels (Fig. 5G). These data suggest that depletion of vigilin impairs DNA DSB repair by the HR repair pathway.
To obtain further insight into the role of vigilin in DNA repair, we analyzed RAD51 recruitment at a single DNA DSB site in control and vigilin-depleted DR95 cells by chromatin immunoprecipitation (ChIP) assay, as described previously (56). Expression of I-SceI generates a DSB at a specific site in the genome, inducing local γ-H2AX formation and subsequent recruitment of repair proteins. We observed significantly decreased RAD51 levels at the damage site after vigilin depletion (Fig. 5H), which suggests that vigilin facilitates the recruitment of signaling and HR-related repair proteins at DNA damage sites. The reduction was not due to decreased RAD51 protein levels since these were similar to those in control cells (Fig. 5I). We used coimmunoprecipitation assays to test if vigilin physically associates with RAD51 or other signaling proteins, such as BRCA1. As shown in Fig. 5J, immunoprecipitated vigilin associated with RAD51 and, conversely, immunoprecipitation of RAD51 confirmed the interaction between vigilin and RAD51 (Fig. 5K). Similar to the RAD51 interaction, we observed interaction between vigilin and BRCA1 (Fig. 5L). These results indicate that vigilin interacts with RAD51 and BRCA1 and facilitates their recruitment to DNA DSB sites.
Ectopic expression restores the DNA damage response in cells depleted of endogenous vigilin.
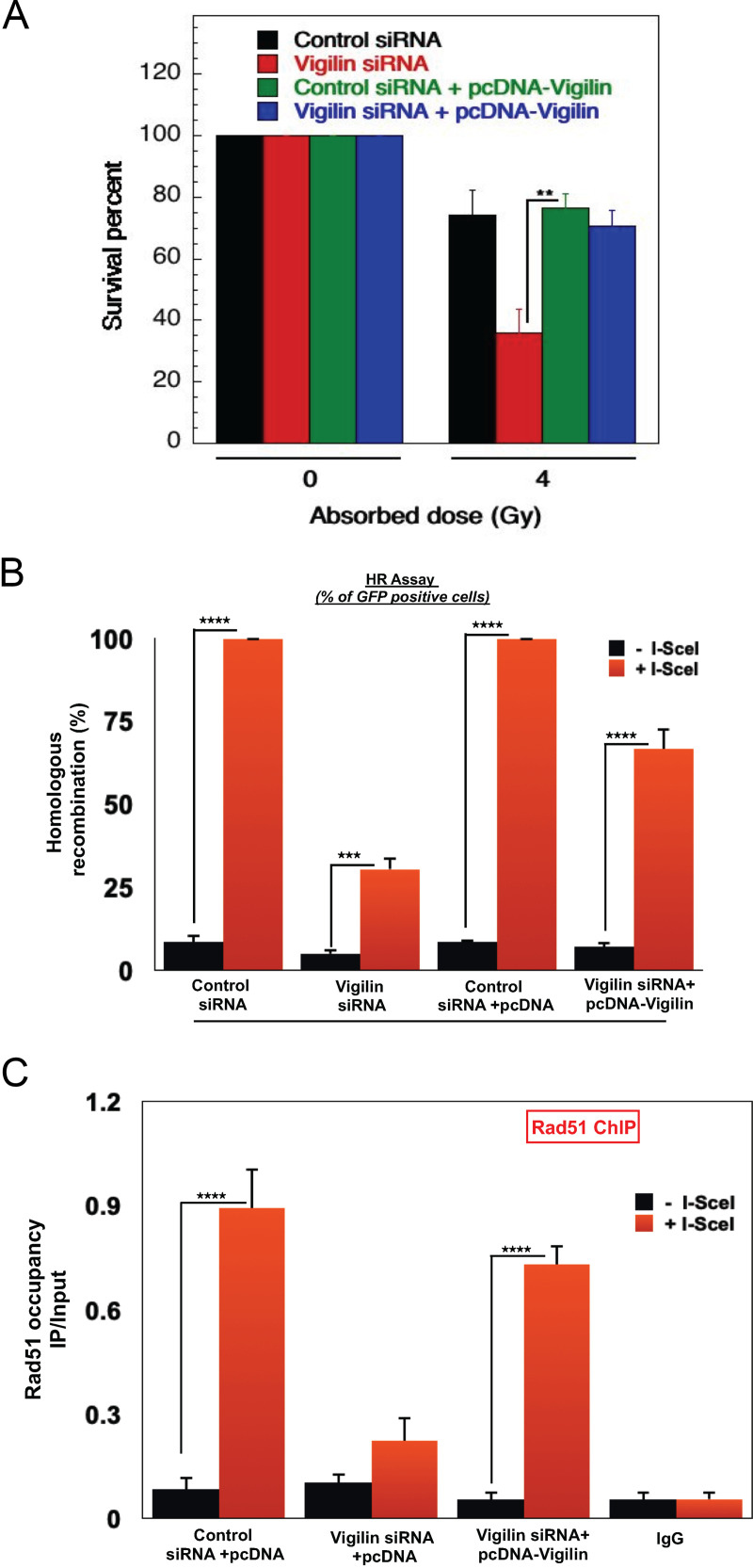
We ectopically expressed pcDNA-vigilin after depletion of endogenous vigilin with untranslated region (UTR)-specific siRNA: we found that ectopic vigilin expression rescued cell survival postirradiation (Fig. 6A). In addition, we measured the reconstitution frequency of the stably integrated GFP-HR reporter gene in control and ectopically expressing vigilin cells. In cells depleted of endogenous vigilin, expression of ectopic vigilin rescued the HR repair defect as the number of GFP-positive cells was significantly increased (Fig. 6B). In line with the rescue phenotypes, we also observed that in cells lacking vigilin, ectopic expression restored RAD51 levels at the DSB sites (Fig. 6C), confirming that vigilin is required for DNA DSB repair by the HR pathway.
FIG 6.
Ectopic expression of vigilin rescues DNA damage repair in vigilin-depleted cells. (A) Clonogenic survival after irradiation of H1299 cells with 4 Gy of IR with and without knockdown of endogenous vigilin and ectopic expression of vigilin. Complementation of vigilin in vigilin-deficient cells rescued cell survival postirradiation. (B) Homologous recombination frequencies in cells are shown after induction of double-strand breaks by I-SecI with and without knockdown of endogenous vigilin and ectopic expression of vigilin. Ectopic vigilin expression in vigilin knockdown cells restores survival and homologous recombination efficiency. (C) Chromatin immunoprecipitation analysis of Rad51 occupancy at the I-SecI site in cells with and without the depletion of vigilin and ectopic expression of vigilin. Ectopic expression of vigilin restores Rad51 occupancy to double-strand breaks in vigilin-deficient cells. The error bars in all experiments represent the standard deviations from three independent experiments.
Acetylation-dependent recruitment of vigilin to DNA damage sites.
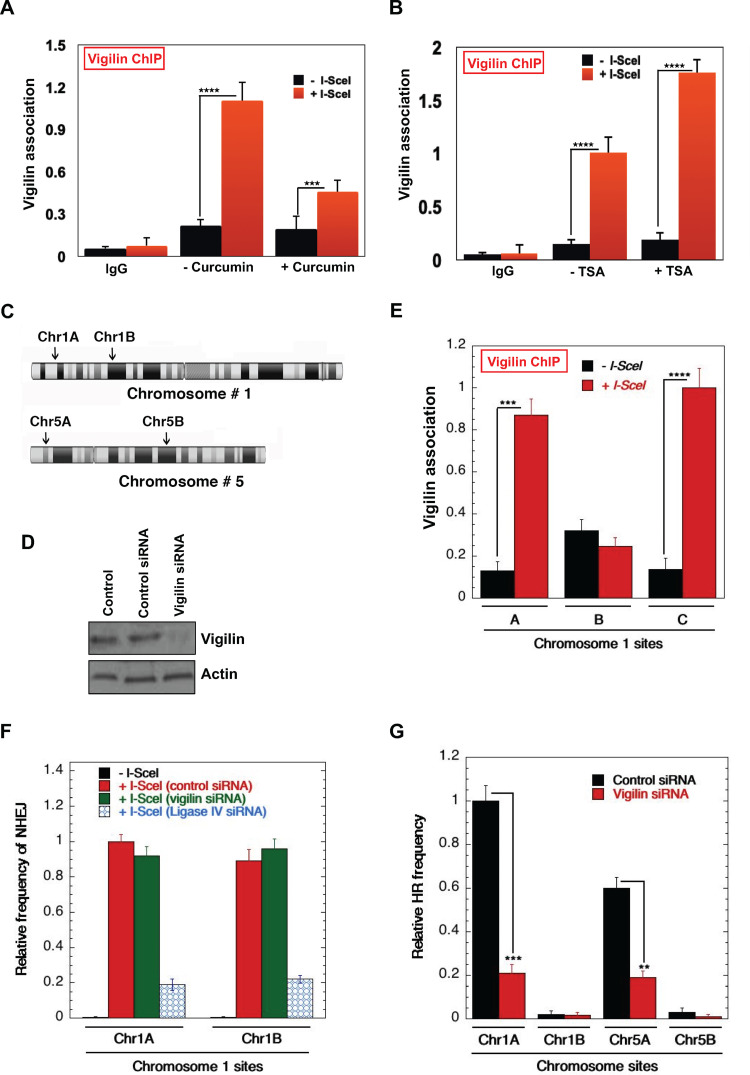
Chromatin modifications have been shown to play important roles in transcription and DNA repair by regulating the recruitment of the requisite factors to chromatin (44–49). We investigated the potential role of histone acetylation in vigilin recruitment by analyzing localization to I-SceI-induced DSBs in control cells and cells treated with curcumin, an inhibitor of the CBP/p300 family of histone acetyltransferases (HATs). Curcumin treatment drastically decreased vigilin recruitment levels compared to untreated cells (Fig. 7A). Furthermore, we also observed increased vigilin association at DSBs in cells treated with trichostatin A (TSA), a histone deacetylase (HDAC) inhibitor (Fig. 7B).
FIG 7.
Acetylation-dependent recruitment of vigilin to DNA damage sites. (A) DR95 cells were transfected with I-SecI plasmid to induce DSB, and cells were treated with 100 μM or left untreated as indicated. Vigilin binding to damage sites was analyzed by chromatin immunoprecipitation in control and curcumin-treated cells. Treatment of cells with the HAT inhibitor curcumin reduces binding of vigilin to DNA damage sites compared to untreated cells. (B) Site-specific breaks were induced by I-SecI, and vigilin’s association with DNA damage sites in control and TSA-treated (20 ng/ml) cells was analyzed by chromatin immunoprecipitation. Treatment of cells with TSA increased binding of vigilin to DNA damage sites compared to that in untreated cells. (C) I-SceI sites were introduced by CRISPR at the indicated positions in gene-rich and gene-poor regions of Chr1 (chromosome 1) and Chr5 (chromosome 5) in H1299 cells using single-stranded oligonucleotides carrying the 18-nucleotide (nt) I-SceI site flanked with 45- to 90-bp homologous sequence (described by Horikoshi et al. [57]). (D) Western blot showing the knockdown of vigilin in H1299 cell lines with site-specific I-SceI sites. (E) Impact of H4K16ac levels on vigilin association with DSB sites (A, Chr1A; B, Chr1B; and C, Chr1C). Vigilin preferentially binds to the regions highly enriched with H4K16 acetylation. (F and G) Site-specific NHEJ and HR repair in control and vigilin-depleted cells, respectively. Chr1A and Chr5A are gene/H4K16ac-rich regions, whereas Chr1B and Chr5B are gene/H4K16ac-poor regions.
We measured vigilin binding to I-SceI-induced DNA DSBs either in transcriptionally active gene-rich and transcriptionally inactive gene-poor regions in control and vigilin-depleted cells (Fig. 7C and D) (57, 58). We found significantly increased binding of vigilin to DSBs in gene-rich regions, which are marked by elevated H4K16ac (chromosome 1A [Chr1A] and Chr1C), compared to a gene-poor region (Chr1B) with reduced H4K16ac marks (Fig. 7E). As shown previously, these regions are well defined as H4K16ac- rich and -poor regions in the genome (57). DNA DSB repair by HR occurs predominately in H4K16ac-containing gene-rich regions of the genome. Since our results detected increased vigilin binding in H4K16ac-rich regions, we examined the impact of vigilin loss on nonhomologous end joining (NHEJ) and HR frequency at I-SceI cleavage sites located in transcriptionally active gene-rich and gene-poor regions. Depletion of vigilin had very little effect on repair by NHEJ (Fig. 7F), whether the repair site was located in a gene-rich region (Chr1A) or gene-poor region (Chr1B). Loss of vigilin, however, significantly impacted HR repair at Chr1A and Chr5A DSB sites, which are in gene- and H4K16ac-rich regions, but had no impact on HR repair frequency at the gene- and H4K16ac-poor Chr1B and Chr5B sites (Fig. 7G). These data support the concept that vigilin binding to DSBs depends upon histone acetylation, consistent with recent findings of a preferential utilization of the HR pathway in gene-rich regions associated with elevated H4K16ac levels (57–59).
Vigilin depletion affects repair of DNA stalled forks.
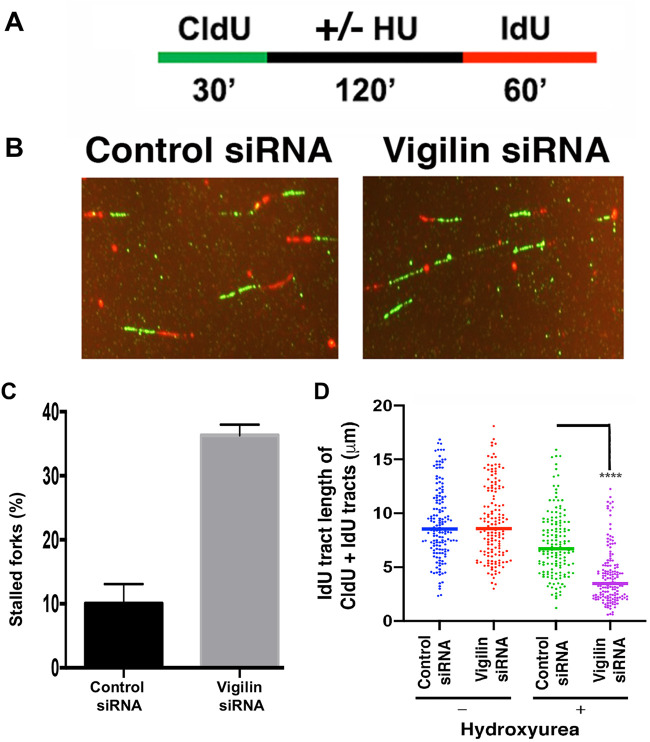
Repair of replicative stress-induced DNA damage utilizes components of the HR repair pathway (60–64). To determine the role of vigilin in stalled fork progression, we used a DNA fiber assay to evaluate replication fork dynamics at a single-molecule resolution in control and vigilin-depleted cells. For these analyses, cells were pulse-labeled with 5-chlorodeoxyuridine (CldU) and then treated with HU for 2 h to induce replication stress by nucleotide pool depletion, followed by HU washout and pulse-labeling with 5-iododeoxyuridine (IdU) (Fig. 8A). After removal of the HU block, vigilin-depleted cells produced fewer contiguous CIdU/ldU signals, and therefore fewer sites that had resumed replication (Fig. 8B and D), indicating defective restart of previously initiated replication origins. Furthermore, in vigilin-depleted cells we observed a higher percentage of stalled DNA replication forks (Fig. 8C). Failure to restart DNA replication at stalled forks indicates a defect in DSB resolution, which is partly dependent on repair by HR.
FIG 8.
Vigilin-depleted cells have defective resolution of stalled DNA replication forks and firing of new origins of replication. (A) Schematic representation of the assay for DNA labeling and HU treatment for the fiber assay. (B) Representative images of new DNA replication forks and reinitiation of stalled DNA replication forks in control and vigilin-depleted cells. (C) Percentage of stalled DNA replication forks in control and vigilin-depleted cells after HU treatment. (D) DNA fiber analysis of IdU track lengths of CldU- plus IdU-containing fibers after hydroxyurea (HU) treatment. Error bars represent standard deviations from three independent experiments.
DISCUSSION
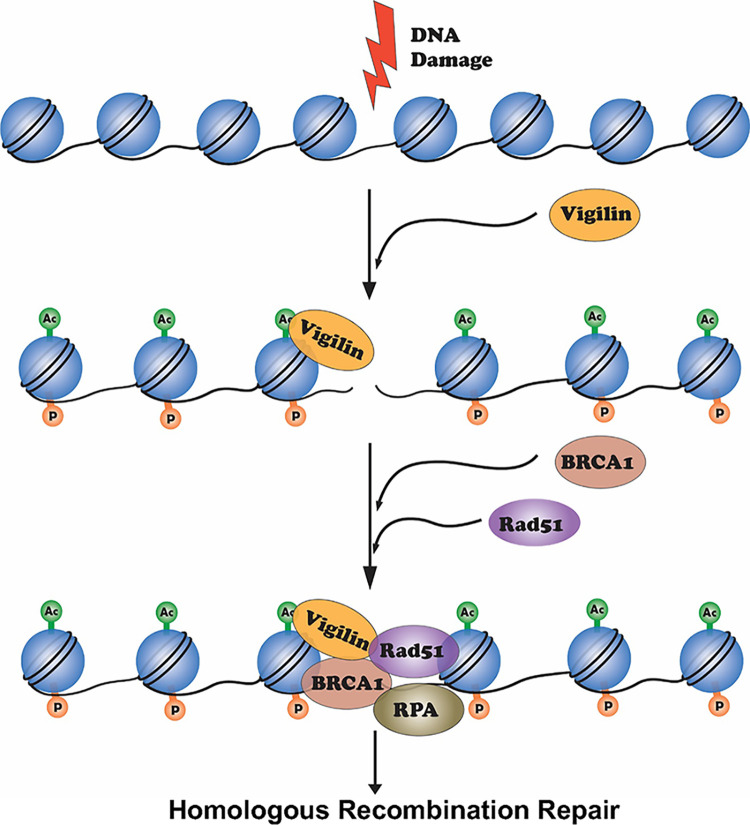
Vigilin is an RNA-binding protein active in a diverse range of biological processes, including heterochromatin formation, chromosome segregation, and mRNA stability (11). Cytoplasmic vigilin is involved in RNA metabolism, while nuclear vigilin regulates heterochromatin structure and chromosome segregation (23, 24). We have reported that Vgl1, the vigilin homologue in S. pombe, is essential for heterochromatin-mediated gene silencing and its loss leads to derepression of heterochromatin at centeromeres and telomers (26). Heterochromatin-associated proteins have been shown to play important roles in DNA repair (65), and we found Vgl1-deficient S. pombe cells were hypersensitive to DNA-damaging agents. Consistent with this role in S. pombe, our data provide strong evidence that vigilin signaling also plays a crucial role in maintenance of mammalian genomic stability. We found that, as in yeast, vigilin interacts with DNA repair factors and depletion of vigilin decreases cell survival postirradiation, likely due to defects in DNA repair (Fig. 1). In the absence of vigilin, cell survival in response to DNA-damaging agents, like ionizing radiation and cisplatin, decreased, while metaphase chromosomal aberrations increased. These abnormalities occurred concomitantly with persistent γ-H2AX, 53BP1, and radiation-induced focus (RIF) DNA repair foci but reduced BRCA1 and RAD51 foci. This implies that vigilin depletion alters pathway choice against error-free homology-directed repair and in favor of error-prone nonhomologous end-joining at sites of DNA DSBs. Furthermore, HR-dependent reconstitution of a nonfunctional interrupted GFP reporter gene was reduced by vigilin depletion but then rescued after its exogenous addition. Vigillin recruitment to DNA-damaged sites was contingent upon histone H4K16 acetylation, as previously observed for other DNA repair factors (66). Importantly, both the number of stalled forks and firing replication origins increased in the absence of vigilin, which suggests a defect in stalled fork resolution due to problems in DSB repair by the HR pathway. In summary upon DNA damage, vigilin is recruited to chromatin in an acetylation-dependent manner and facilitates recruitment of RAD51/BRCA1 to damaged sites, which then promotes homology-directed DNA repair (Fig. 9).
FIG 9.
Proposed model for the role of vigilin in DNA repair. Upon DNA damage, H2AX is phosphorylated on serine 139 by ATM/ATR kinases. This phosphorylation creates a binding site for various proteins like 53BP1, which affects its downstream target, Rif1. Vigilin in response to DNA damage gets recruited to DNA damage sites in an acetylation-dependent manner and facilitates the recruitment of Rad51/BRCA1 to DNA DSB sites to replace 53BP1 for facilitation of MRN-mediated DNA end resection by RPA loading onto the resulting ssDNA and polymerization by RAD51.
Our results uncover a novel role for vigilin in DNA DSB repair with implications in autism-related disorders, where vigilin has been shown to be significantly downregulated and associated with accumulated DSBs. Fibroblasts from ASD-affected infants and toddlers reprogrammed into neural progenitor cells (NPCs) display increased cell proliferation (67). These NPCs also exhibit elevated numbers of DSBs, particularly in extremely long genes with critical roles in adherens junctions, apical polarity, and cell migration, thereby reducing their overall expression (68) and limiting excitatory synapses. Biochemically, ASD-derived NPCs display spontaneously increased γ-H2AX foci, shorter origin-to-origin replication firing, chronic activation of the ATR-CHK1 pathway, and elevated DNA damage (68). It has been hypothesized that haploinsufficiency of vigilin may be a contributing factor in ASD-related phenotypes (1). Therefore, given that, as we show, vigilin plays a key role in DNA repair processes, it now appears important to elucidate the mechanism by which vigilin contributes to the conflicts between transcription and replication that are not resolved due to defects in DNA repair that may be related to ASD pathogenesis.
Our findings that vigilin is a DNA repair factor have key implications for its role in various cancers with which it has been shown to be associated. Vigilin is found to be upregulated in ovarian cancer cells, gastric cancer, and leukemic and prostate cancer cells (69–72). In breast cancer cells, vigilin competes with the RNA-binding protein HuR for c-fms proto-oncogene mRNA, thereby promoting its degradation. Vigilin overexpression decreases the half-life of c-fms mRNA and suppresses its translation (6). Overexpression of c-fms in breast cancers confers invasiveness and metastatic properties, suggesting that degradation of c-fms RNA by vigilin is an important regulator of c-fms and may function in tumor repression. Vigilin has also been shown to be overexpressed in human hepatocellular carcinomas. The level of vigilin overexpression correlates with benign lesions, liver cirrhosis, and hepatocellular carcinoma (HCC). Vigilin has been suggested to play a crucial role in HCC cell proliferation, survival, migration, and tumor growth (73). It will be important to understand the contribution of vigilin-mediated DNA repair processes to these cancers. Our results are also in line with the observation that activation of oncogenes like c-Myc drives cellular proliferation and induces chromosomal abnormalities. Genomic instability is considered one of the hallmarks of cancer, generating mutations that drive various cancers (33). Defects affecting various DNA repair factors known as caretakers of the genome have been implicated in cancer progression (74). Furthermore, reduced mutational load in transcriptionally active open chromatin sites depends upon efficient targeting of oxidized DNA base repair to these sites (75). Our finding here that vigilin promotes DSB repair by HR in transcriptionally active gene-rich chromatin sites that is impaired by its dysregulation therefore has key implications for mechanisms underlying both ASD pathogenesis and error-prone proliferation in cancers.
Our results are also in line with several reports of RNA-binding proteins (RBPs) being involved in the DNA damage response through multiple mechanisms, including control of R loop formation and regulation of the selective expression of DDR genes upon DNA damage (76). Our results show that vigilin directly binds to DNA damage sites and interacts with DNA repair proteins to control their recruitment to damage sites (77). Several key DNA repair proteins like Ku70 to Ku80 heterodimer, DNA-protein kinase (PK), BRCA1, and 53BP1 have been shown to directly interact with RNAs and regulate their repair efficiency (77–80). It would be critical to assess whether the function of vigilin in DNA repair depends on an interaction with RNA. If vigilin’s role in DNA repair is not independent of RNA, it will be interesting in the future to dissect the role of the vigilin RNA binding domains in DNA repair and identify the repertoire of RNAs required for its function.
In summary, our results have added vigilin to the list of RNA-binding proteins having a role in DNA double-strand repair and uncover a novel role in the DNA damage response that could be a contributing factor in vigilin’s role in various cancers and other human diseases.
MATERIALS AND METHODS
Cell lines and transfection.
HeLa and H1299 cell lines containing a stably integrated DR-GFP gene cassette were maintained in Dulbecco’s modified Eagle’s medium (DMEM)–F-12 medium (D5671; Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal bovine serum (F2442; Sigma-Aldrich) and 1% penicillin–streptomycin. Small interfering RNAs (siRNAs) for vigilin and control siRNAs or short hairpin RNAs (shRNAs) were purchased from Dharmacon Research (Lafayette, CO). Cells were transfected with either control siRNA or vigilin siRNA/shRNA using an Amaxa Nucleofector according to the manufacturer's protocol (Lonza, Allendale, NJ)
Clonogenic assay.
After transfection with either control or vigilin siRNA, 60-mm dishes were seeded with 600 HeLa or H1299 cells in triplicate. The cells were irradiated using a Rad Source RS 2000 X-ray irradiator (Rad Source Technologies, Inc., GA). Six-well dishes were seeded with different numbers of cells transfected with either vigilin siRNA or control siRNA with or without radiation. For replication stress, cells were treated with indicated concentrations of cisplatin for 1 h and then washed three times with phosphate-buffered saline (PBS) and refed with fresh DMEM. The treated and control cells were cultured for 14 days, and the colonies were fixed and then stained with crystal violet. Colonies with more than 50 cells were scored, and results were normalized for the plating efficiency of each cell line as described earlier (81).
Chromosomal aberration analysis.
Specific chromosomal aberrations were analyzed in metaphase cells by a procedure described earlier (30). Chromosome aberrations were assessed after exponentially growing control and vigilin-depleted cells were irradiated with 2 Gy of IR, and metaphase cells were processed 4 to 5 h after IR, as previously described (30). Cells were then treated with colcemid for 3 h, followed by hypotonic treatment (0.56% [wt/vol] KCl) at room temperature. Cells were fixed with fresh methanol-acetic acid (3:1) before scoring of chromosome aberrations by Giemsa staining (4% in 0.01 M phosphate buffer [pH 6.8]) for 10 min.
Telomeric FISH.
Telomeres in metaphase spreads were detected by fluorescent in situ hybridization (FISH) with a telomere-specific probe as described previously (30, 82). To detect chromosomal associations, we used centromere and telomere probe combinations. Centromere-specific peptide nucleic acid (PNA) probes were labeled with fluorescein isothiocyanate (FITC; green), and telomere-specific PNA probes were labeled with the fluorochrome Cy3 (red).
DR-GFP assay.
The homologous recombination assay was performed as described previously (81, 83) using a direct repeat-green fluorescent protein (DR-GFP) cassette, which is stably integrated in H1299 cells. Briefly, H1299 cells were transfected with control siRNA or vigilin siRNA and grown for 48 h prior to a second transfection with an I-SceI expression plasmid using a Nucleofector 2b device. Cells were trypsinized 48 h after transfection, and the percentage of GFP-positive cells after I-Scel DSB induction was determined by flow cytometry for measurement of HR repair. Site-specific DNA HR and NHEJ assays in gene- and H4K16ac-rich and -poor regions were performed as described previously (57).
Chromatin immunoprecipitation assay.
Chromatin immunoprecipitation assays (ChIP) were performed as described previously (56, 57, 83, 84). Briefly, DR95 cells carrying a stably integrated I-SceI site were transfected with I-Scel expression plasmid after a previous transfection with control siRNA or vigilin siRNA. Cells were cross-linked with 1% formaldehyde for 10 min with gentle shaking at room temperature followed by quenching using 125 mM glycine for 5 min. Cells were washed with ice-cold PBS and resuspended in 300 μl of sonication buffer (50 mM Tris-HCl [pH 8], 10 mM EDTA, 1% SDS, and protease inhibitors). Cross-linked chromatin was sonicated with the Diagenode Bioruptor to yield DNA fragments of an average size of 200 to 600 bp. Immunoprecipitations were carried out with Rad51/vigilin antibodies. The primers used in the PCR were first analyzed for both efficiency and linear range by real-time PCR with a LightCycler (Applied Biosystems). All ChIP experiments were performed three times and yielded consistent results. The results shown are based on three independent experiments with standard errors.
Immunofluorescence microscopy.
For immunofluorescence, H1299 cells were transfected with control siRNA or vigilin siRNA, grown for 48 h, and then irradiated and allowed to recover for the indicated time intervals (27, 41, 81). For immunostaining, cells were fixed in 4% paraformaldehyde for 5 min at room temperature, permeabilized with 1× PBS containing 0.5% Triton X-100 for 5 min, blocked in PBS with 1% bovine serum albumin, and washed twice with 1× PBS followed by incubation with the desired primary antibody (γ-H2AX or 53BP1) for 30 min at room temperature. After a subsequent incubation with secondary antibody (Alexa Fluor 488-conjugated goat anti-rabbit/mouse secondary antibody), fluorescent images of foci were captured with a Zeiss Axio Imager M2 microscope, and the foci were counted by ImageJ software. The results shown are from three independent experiments.
DNA replication restart assay.
DNA fiber assays were performed to assess replication fork stalling, and fork protection as described previously (55, 85). Cells were pulse-labeled with CldU (100 μM, 30 min) and replaced by medium with or without the HU and followed by IdU (150 μM, 60 min). DNA fibers were spread by gravity and stained using immunofluorescent procedures with CldU- and IdU-specific antibodies. Replication recovery was assessed by measuring the tract length of the IdU, with or without HU treatment of tracts that contain both CldU and IdU labels. DNA fibers were imaged by using a 1041 Carl Zeiss Axio Imager D2 microscope. Statistical analysis was performed using the Mann-Whitney test.
DNA end resection assay.
A quantitative DNA resection assay was used to measure ssDNA as described previously (56, 60), and developmental details of the ER-AsiSI-based resection assay are the same as described previously (61). Briefly, ssDNA was quantified using the hemagglutinin (HA)-ER-AsiSI system in which AsiSI enzyme is fused with a hormone-binding domain (HA-ER-AsiSI) and stably expressed in U2OS cells. Addition of 4-hydroxy-tamoxifen (4-OHT) relocalizes the AsiSI enzyme to the nucleus, which generates a double-strand break at specific AsiSI sequence (GCGATCGC) sites. U2OS cells (6 × 106 cells per ml) transfected with either control siRNA, vigilin siRNA, or Mre11 siRNA were mixed with 0.6% low-melting-temperature agarose in PBS. Fifty microliters of cell suspension was solidified as an agar ball by dropping the suspension on Parafilm. The agar ball was treated serially with 1 ml of ESP buffer containing 2% N-lauroylsarcosine, 0.5 M EDTA, 1 mM CaCl2 (pH 8.0), and 1 mg/ml of proteinase K and then with high-salt (HS) buffer containing 0.15 M KCl, 2 mM EDTA, 5 mM MgCl2, 1.85 M NaCl, 0.5% Triton X-100 (pH 7.5), and 4 mM Tris at 16°C with rotation for 20 h. The sample was then washed with 1 ml of phosphate buffer containing 0.8 mM MgCl2 (pH 7.4), 133 mM KCl, 1.5 mM KH2PO4, and 8 mM Na2HPO4 for 1 h at 4°C with rotation. After heating the agar ball at 70°C for 10 min, the samples were diluted 15-fold with 70°C double-distilled water and mixed with 2× NEB restriction enzyme buffer. Genomic DNA samples were digested with 60 U of restriction enzyme BsrGI (New England BioLabs) or mock digested at 37°C overnight. BsrGI-digested genomic DNA samples were used as the templates to score ssDNA at the specific AsiSI sites by quantitative PCR (qPCR).
Immunoprecipitations.
Exponentially growing cells were lysed in ice-cold lysis buffer (50 mM Tris-HCl [pH 7.4], 200 mM sodium chloride, 2 mM EDTA, 0.5% Na-deoxycholate, 1% NP-48) containing protease inhibitors, aprotinin (0.5 mg/ml), leupeptin (0.5 mg/ml), phenylmethylsulfonyl fluoride (PMSF; 1 mM), sodium orthovanadate (0.2 mM), and sodium fluoride (50 mM). Protein concentrations were normalized using the Bio-Rad protein assay, and the supernatant was incubated with vigilin antibody (Abcam) overnight at 4°C. IgG-conjugated Dynabeads were added the next day and incubated at 4°C for 2 h. Immunoprecipitated material was washed four times with lysis buffer, resuspended in SDS sample buffer, and analyzed by SDS-PAGE followed by Western blotting using RAD51 vigilin and BRCA1 antibodies.
Data availability.
This study includes no data deposited in external repositories.
ACKNOWLEDGMENTS
We are highly grateful to the members of the Pandita laboratory for their support during the execution of present work. We acknowledge Ishfaq Ahamd Pandith for help in the preparation of the working model.
The work was supported by grants from Department of Biotechnology, Government of India (BT/PR16122/BRB/10/1478/2016 and BT/PR22348/BRB/10/1625/2017), DST-SERB (EMR/2016/000958 and CRG/2020/003632) Government of India, and an ICMR Fellowship for Young Biomedical Scientists [no. INDO/FRC/452/(Y-53)/2016-17-IHD, made available through Houston Methodist Research Institute, Houston, TX] to M.A. T.K.P. acknowledges funds from Houston Methodist Research Institute, M. D. Anderson Cancer Center, Houston, and National Institutes of Health grants CA129537 and GM109768. J.A.T. is supported by Cancer Prevention Research Institute of Texas (CPRIT) grant RP180813, by NIH grants PO1 CA092584 and R35 CA220430, and by a Robert A. Welch Chemistry Chair.
S.B., R.K.P, A.M., A.B., U.S.M., D.K.S., S.J., K.P.B., V.K.C., and M.A. carried out the experiments. S.B., R.K.P., A.M., A.B., J.A.T., T.K.P., and M.A. designed the experiments and interpreted the results. S.B., R.K.P., A.M., A.B., C.R.H., G.R. J.A.T., T.K.P., and M.A. wrote the manuscript, and all the authors edited and approved it.
We declare we have no conflicts of interest.
Contributor Information
Tej K. Pandita, Email: tej.pandita@bcm.edu.
Mohammad Altaf, Email: altafbhat@uok.edu.in.
REFERENCES
- 1.Felder B, Radlwimmer B, Benner A, Mincheva A, Todt G, Beyer KS, Schuster C, Bolte S, Schmotzer G, Klauck SM, Poustka F, Lichter P, Poustka A. 2009. FARP2, HDLBP and PASK are downregulated in a patient with autism and 2q37.3 deletion syndrome. Am J Med Genet 149A:952–959. 10.1002/ajmg.a.32779. [DOI] [PubMed] [Google Scholar]
- 2.O'Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, Carvill G, Kumar A, Lee C, Ankenman K, Munson J, Hiatt JB, Turner EH, Levy R, O'Day DR, Krumm N, Coe BP, Martin BK, Borenstein E, Nickerson DA, Mefford HC, Doherty D, Akey JM, Bernier R, Eichler EE, Shendure J. 2012. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338:1619–1622. 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, Turner EH, Stanaway IB, Vernot B, Malig M, Baker C, Reilly B, Akey JM, Borenstein E, Rieder MJ, Nickerson DA, Bernier R, Shendure J, Eichler EE. 2012. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485:246–250. 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kosmicki JA, Samocha KE, Howrigan DP, Sanders SJ, Slowikowski K, Lek M, Karczewski KJ, Cutler DJ, Devlin B, Roeder K, Buxbaum JD, Neale BM, MacArthur DG, Wall DP, Robinson EB, Daly MJ. 2017. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat Genet 49:504–510. 10.1038/ng.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, Smith JD, Paeper B, Nickerson DA, Dea J, Dong S, Gonzalez LE, Mandell JD, Mane SM, Murtha MT, Sullivan CA, Walker MF, Waqar Z, Wei L, Willsey AJ, Yamrom B, Lee YH, Grabowska E, Dalkic E, Wang Z, Marks S, Andrews P, Leotta A, Kendall J, Hakker I, Rosenbaum J, Ma B, Rodgers L, Troge J, Narzisi G, Yoon S, Schatz MC, Ye K, McCombie WR, Shendure J, Eichler EE, State MW, Wigler M. 2014. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515:216–221. 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woo HH, Yi X, Lamb T, Menzl I, Baker T, Shapiro DJ, Chambers SK. 2011. Posttranscriptional suppression of proto-oncogene c-fms expression by vigilin in breast cancer. Mol Cell Biol 31:215–225. 10.1128/MCB.01031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Courchet V, Roberts AJ, Meyer-Dilhet G, Del Carmine P, Lewis TL, Jr, Polleux F, Courchet J. 2018. Haploinsufficiency of autism spectrum disorder candidate gene NUAK1 impairs cortical development and behavior in mice. Nat Commun 9:4289. 10.1038/s41467-018-06584-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nuytens K, Gantois I, Stijnen P, Iscru E, Laeremans A, Serneels L, Van Eylen L, Liebhaber SA, Devriendt K, Balschun D, Arckens L, Creemers JW, D'Hooge R. 2013. Haploinsufficiency of the autism candidate gene Neurobeachin induces autism-like behaviors and affects cellular and molecular processes of synaptic plasticity in mice. Neurobiol Dis 51:144–151. 10.1016/j.nbd.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Wang S, Tan N, Zhu X, Yao M, Wang Y, Zhang X, Xu Z. 2018. Sh3rf2 haploinsufficiency leads to unilateral neuronal development deficits and autistic-like behaviors in mice. Cell Rep 25:2963–2971.e6. 10.1016/j.celrep.2018.11.044. [DOI] [PubMed] [Google Scholar]
- 10.Bagni C, Zukin RS. 2019. A synaptic perspective of fragile X syndrome and autism spectrum disorders. Neuron 101:1070–1088. 10.1016/j.neuron.2019.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng MH, Jansen RP. 2017. A jack of all trades: the RNA-binding protein vigilin. Wiley Interdiscip Rev RNA 8:e1448. 10.1002/wrna.1448. [DOI] [PubMed] [Google Scholar]
- 12.Burd CG, Dreyfuss G. 1994. Conserved structures and diversity of functions of RNA-binding proteins. Science 265:615–621. 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 13.Kugler S, Grunweller A, Probst C, Klinger M, Muller PK, Kruse C. 1996. Vigilin contains a functional nuclear localisation sequence and is present in both the cytoplasm and the nucleus. FEBS Lett 382:330–334. 10.1016/0014-5793(96)00204-9. [DOI] [PubMed] [Google Scholar]
- 14.Kanamori H, Dodson RE, Shapiro DJ. 1998. In vitro genetic analysis of the RNA binding site of vigilin, a multi-KH-domain protein. Mol Cell Biol 18:3991–4003. 10.1128/MCB.18.7.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dodson RE, Shapiro DJ. 1997. Vigilin, a ubiquitous protein with 14 K homology domains, is the estrogen-inducible vitellogenin mRNA 3'-untranslated region-binding protein. J Biol Chem 272:12249–12252. 10.1074/jbc.272.19.12249. [DOI] [PubMed] [Google Scholar]
- 16.Weber V, Wernitznig A, Hager G, Harata M, Frank P, Wintersberger U. 1997. Purification and nucleic-acid-binding properties of a Saccharomyces cerevisiae protein involved in the control of ploidy. Eur J Biochem 249:309–317. 10.1111/j.1432-1033.1997.00309.x. [DOI] [PubMed] [Google Scholar]
- 17.Wintersberger U, Kuhne C, Karwan A. 1995. Scp160p, a new yeast protein associated with the nuclear membrane and the endoplasmic reticulum, is necessary for maintenance of exact ploidy. Yeast 11:929–944. 10.1002/yea.320111004. [DOI] [PubMed] [Google Scholar]
- 18.Hirschmann WD, Westendorf H, Mayer A, Cannarozzi G, Cramer P, Jansen RP. 2014. Scp160p is required for translational efficiency of codon-optimized mRNAs in yeast. Nucleic Acids Res 42:4043–4055. 10.1093/nar/gkt1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cunningham KS, Dodson RE, Nagel MA, Shapiro DJ, Schoenberg DR. 2000. Vigilin binding selectively inhibits cleavage of the vitellogenin mRNA 3'-untranslated region by the mRNA endonuclease polysomal ribonuclease 1. Proc Natl Acad Sci U S A 97:12498–12502. 10.1073/pnas.220425497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Q, Yang B, Xie X, Wei L, Liu W, Yang W, Ge Y, Zhu Q, Zhang J, Jiang L, Yu X, Shen W, Li R, Shi X, Li B, Qin Y. 2014. Vigilin interacts with CCCTC-binding factor (CTCF) and is involved in CTCF-dependent regulation of the imprinted genes Igf2 and H19. FEBS J 281:2713–2725. 10.1111/febs.12816. [DOI] [PubMed] [Google Scholar]
- 21.Wilson LC, Leverton K, Oude Luttikhuis ME, Oley CA, Flint J, Wolstenholme J, Duckett DP, Barrow MA, Leonard JV, Read AP. 1995. Brachydactyly and mental retardation: an Albright hereditary osteodystrophy-like syndrome localized to 2q37. Am J Hum Genet 56:400–407. [PMC free article] [PubMed] [Google Scholar]
- 22.Phelan MC, Rogers RC, Clarkson KB, Bowyer FP, Levine MA, Estabrooks LL, Severson MC, Dobyns WB. 1995. Albright hereditary osteodystrophy and del(2) (q37.3) in four unrelated individuals. Am J Med Genet 58:1–7. 10.1002/ajmg.1320580102. [DOI] [PubMed] [Google Scholar]
- 23.Huertas D, Cortes A, Casanova J, Azorin F. 2004. Drosophila DDP1, a multi-KH-domain protein, contributes to centromeric silencing and chromosome segregation. Curr Biol 14:1611–1620. 10.1016/j.cub.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 24.Zhou J, Wang Q, Chen LL, Carmichael GG. 2008. On the mechanism of induction of heterochromatin by the RNA-binding protein vigilin. RNA 14:1773–1781. 10.1261/rna.1036308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cortes A, Huertas D, Fanti L, Pimpinelli S, Marsellach FX, Pina B, Azorin F. 1999. DDP1, a single-stranded nucleic acid-binding protein of Drosophila, associates with pericentric heterochromatin and is functionally homologous to the yeast Scp160p, which is involved in the control of cell ploidy. EMBO J 18:3820–3833. 10.1093/emboj/18.13.3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farooq Z, Abdullah E, Banday S, Ganai SA, Rashid R, Mushtaq A, Rashid S, Altaf M. 2019. Vigilin protein Vgl1 is required for heterochromatin-mediated gene silencing in Schizosaccharomyces pombe. J Biol Chem 294:18029–18040. 10.1074/jbc.RA119.009262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dinant C, Luijsterburg MS. 2009. The emerging role of HP1 in the DNA damage response. Mol Cell Biol 29:6335–6340. 10.1128/MCB.01048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma GG, Hwang KK, Pandita RK, Gupta A, Dhar S, Parenteau J, Agarwal M, Worman HJ, Wellinger RJ, Pandita TK. 2003. Human heterochromatin protein 1 isoforms HP1(Hsalpha) and HP1(Hsbeta) interfere with hTERT-telomere interactions and correlate with changes in cell growth and response to ionizing radiation. Mol Cell Biol 23:8363–8376. 10.1128/MCB.23.22.8363-8376.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luijsterburg MS, Dinant C, Lans H, Stap J, Wiernasz E, Lagerwerf S, Warmerdam DO, Lindh M, Brink MC, Dobrucki JW, Aten JA, Fousteri MI, Jansen G, Dantuma NP, Vermeulen W, Mullenders LH, Houtsmuller AB, Verschure PJ, van Driel R. 2009. Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol 185:577–586. 10.1083/jcb.200810035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pandita RK, Sharma GG, Laszlo A, Hopkins KM, Davey S, Chakhparonian M, Gupta A, Wellinger RJ, Zhang J, Powell SN, Roti Roti JL, Lieberman HB, Pandita TK. 2006. Mammalian Rad9 plays a role in telomere stability, S- and G2-phase-specific cell survival, and homologous recombinational repair. Mol Cell Biol 26:1850–1864. 10.1128/MCB.26.5.1850-1864.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Charaka V, Tiwari A, Pandita RK, Hunt CR, Pandita TK. 2020. Role of HP1beta during spermatogenesis and DNA replication. Chromosoma 129:215–226. 10.1007/s00412-020-00739-4. [DOI] [PubMed] [Google Scholar]
- 32.Negrini S, Gorgoulis VG, Halazonetis TD. 2010. Genomic instability—an evolving hallmark of cancer. Nat Rev Mol Cell Biol 11:220–228. 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 33.Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144:646–674. 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 34.Cann KL, Dellaire G. 2011. Heterochromatin and the DNA damage response: the need to relax. Biochem Cell Biol 89:45–60. 10.1139/O10-113. [DOI] [PubMed] [Google Scholar]
- 35.Agarwal M, Pandita S, Hunt CR, Gupta A, Yue X, Khan S, Pandita RK, Pratt D, Shay JW, Taylor JS, Pandita TK. 2008. Inhibition of telomerase activity enhances hyperthermia-mediated radiosensitization. Cancer Res 68:3370–3378. 10.1158/0008-5472.CAN-07-5831. [DOI] [PubMed] [Google Scholar]
- 36.Pandita TK. 2001. The role of ATM in telomere structure and function. Radiat Res 156:642–647.2.0.CO;2]. 10.1667/0033-7587(2001)156[0642:TROAIT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 37.Pandita TK. 2002. ATM function and telomere stability. Oncogene 21:611–618. 10.1038/sj.onc.1205060. [DOI] [PubMed] [Google Scholar]
- 38.Deng Y, Chan SS, Chang S. 2008. Telomere dysfunction and tumour suppression: the senescence connection. Nat Rev Cancer 8:450–458. 10.1038/nrc2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prescott J, Wentzensen IM, Savage SA, De Vivo I. 2012. Epidemiologic evidence for a role of telomere dysfunction in cancer etiology. Mutat Res 730:75–84. 10.1016/j.mrfmmm.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pandita TK, Geard CR. 1996. Chromosome aberrations in human fibroblasts induced by monoenergetic neutrons. I. Relative biological effectiveness. Radiat Res 145:730–739. 10.2307/3579364. [DOI] [PubMed] [Google Scholar]
- 41.Gupta A, Sharma GG, Young CS, Agarwal M, Smith ER, Paull TT, Lucchesi JC, Khanna KK, Ludwig T, Pandita TK. 2005. Involvement of human MOF in ATM function. Mol Cell Biol 25:5292–5305. 10.1128/MCB.25.12.5292-5305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunt CR, Dix DJ, Sharma GG, Pandita RK, Gupta A, Funk M, Pandita TK. 2004. Genomic instability and enhanced radiosensitivity in Hsp70.1- and Hsp70.3-deficient mice. Mol Cell Biol 24:899–911. 10.1128/MCB.24.2.899-911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smilenov LB, Dhar S, Pandita TK. 1999. Altered telomere nuclear matrix interactions and nucleosomal periodicity in ataxia telangiectasia cells before and after ionizing radiation treatment. Mol Cell Biol 19:6963–6971. 10.1128/MCB.19.10.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 10:886–895. 10.1016/S0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 45.Pandita TK, Richardson C. 2009. Chromatin remodeling finds its place in the DNA double-strand break response. Nucleic Acids Res 37:1363–1377. 10.1093/nar/gkn1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott SP, Pandita TK. 2006. The cellular control of DNA double-strand breaks. J Cell Biochem 99:1463–1475. 10.1002/jcb.21067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hunt CR, Ramnarain D, Horikoshi N, Iyengar P, Pandita RK, Shay JW, Pandita TK. 2013. Histone modifications and DNA double-strand break repair after exposure to ionizing radiations. Radiat Res 179:383–392. 10.1667/RR3308.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altaf M, Saksouk N, Cote J. 2007. Histone modifications in response to DNA damage. Mutat Res 618:81–90. 10.1016/j.mrfmmm.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 49.Altaf M, Auger A, Covic M, Cote J. 2009. Connection between histone H2A variants and chromatin remodeling complexes. Biochem Cell Biol 87:35–50. 10.1139/O08-140. [DOI] [PubMed] [Google Scholar]
- 50.Callen E, Di Virgilio M, Kruhlak MJ, Nieto-Soler M, Wong N, Chen HT, Faryabi RB, Polato F, Santos M, Starnes LM, Wesemann DR, Lee JE, Tubbs A, Sleckman BP, Daniel JA, Ge K, Alt FW, Fernandez-Capetillo O, Nussenzweig MC, Nussenzweig A. 2013. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell 153:1266–1280. 10.1016/j.cell.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daley JM, Sung P. 2014. 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol Cell Biol 34:1380–1388. 10.1128/MCB.01639-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Callen E, Zong D, Wu W, Wong N, Stanlie A, Ishikawa M, Pavani R, Dumitrache LC, Byrum AK, Mendez-Dorantes C, Martinez P, Canela A, Maman Y, Day A, Kruhlak MJ, Blasco MA, Stark JM, Mosammaparast N, McKinnon PJ, Nussenzweig A. 2020. 53BP1 enforces distinct pre- and post-resection blocks on homologous recombination. Mol Cell 77:26–38.e7. 10.1016/j.molcel.2019.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jasin M. 2002. Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene 21:8981–8993. 10.1038/sj.onc.1206176. [DOI] [PubMed] [Google Scholar]
- 54.Jasin M, Rothstein R. 2013. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol 5:a012740. 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh DK, Pandita RK, Singh M, Chakraborty S, Hambarde S, Ramnarain D, Charaka V, Ahmed KM, Hunt CR, Pandita TK. 2018. MOF suppresses replication stress and contributes to resolution of stalled replication forks. Mol Cell Biol 38:e00484-17. 10.1128/MCB.00484-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chakraborty S, Pandita RK, Hambarde S, Mattoo AR, Charaka V, Ahmed KM, Iyer SP, Hunt CR, Pandita TK. 2018. SMARCAD1 phosphorylation and ubiquitination are required for resection during DNA double-strand break repair. iScience 2:123–135. 10.1016/j.isci.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Horikoshi N, Sharma D, Leonard F, Pandita RK, Charaka VK, Hambarde S, Horikoshi NT, Gaur Khaitan P, Chakraborty S, Cote J, Godin B, Hunt CR, Pandita TK. 2019. Pre-existing H4K16ac levels in euchromatin drive DNA repair by homologous recombination in S-phase. Commun Biol 2:253. 10.1038/s42003-019-0498-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hunt CR, Pandita TK. 2019. “What’s past is prologue”: pre-existing epigenetic transcriptional marks may also influence DNA repair pathway choice. Radiat Res 192:577–578. 10.1667/RR15541.1. [DOI] [PubMed] [Google Scholar]
- 59.Sharma GG, So S, Gupta A, Kumar R, Cayrou C, Avvakumov N, Bhadra U, Pandita RK, Porteus MH, Chen DJ, Cote J, Pandita TK. 2010. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol 30:3582–3595. 10.1128/MCB.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xie M, Park D, You S, Li R, Owonikoko TK, Wang Y, Doetsch PW, Deng X. 2015. Bcl2 inhibits recruitment of Mre11 complex to DNA double-strand breaks in response to high-linear energy transfer radiation. Nucleic Acids Res 43:960–972. 10.1093/nar/gku1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou Y, Caron P, Legube G, Paull TT. 2014. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res 42:e19. 10.1093/nar/gkt1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rodrigue A, Lafrance M, Gauthier MC, McDonald D, Hendzel M, West SC, Jasin M, Masson JY. 2006. Interplay between human DNA repair proteins at a unique double-strand break in vivo. EMBO J 25:222–231. 10.1038/sj.emboj.7600914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pierce AJ, Johnson RD, Thompson LH, Jasin M. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 13:2633–2638. 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. 2011. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145:529–542. 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bártová E, Malyšková B, Komůrková D, Legartová S, Suchánková J, Krejčí J, Kozubek S. 2017. Function of heterochromatin protein 1 during DNA repair. Protoplasma 254:1233–1240. 10.1007/s00709-017-1090-3. [DOI] [PubMed] [Google Scholar]
- 66.Narita T, Weinert BT, Choudhary C. 2019. Functions and mechanisms of non-histone protein acetylation. Nat Rev Mol Cell Biol 20:156–174. 10.1038/s41580-018-0081-3. [DOI] [PubMed] [Google Scholar]
- 67.Marchetto MC, Belinson H, Tian Y, Freitas BC, Fu C, Vadodaria K, Beltrao-Braga P, Trujillo CA, Mendes APD, Padmanabhan K, Nunez Y, Ou J, Ghosh H, Wright R, Brennand K, Pierce K, Eichenfield L, Pramparo T, Eyler L, Barnes CC, Courchesne E, Geschwind DH, Gage FH, Wynshaw-Boris A, Muotri AR. 2017. Altered proliferation and networks in neural cells derived from idiopathic autistic individuals. Mol Psychiatry 22:820–835. 10.1038/mp.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang M, Wei PC, Lim CK, Gallina IS, Marshall S, Marchetto MC, Alt FW, Gage FH. 2020. Increased neural progenitor proliferation in a hiPSC model of autism induces replication stress-associated genome instability. Cell Stem Cell 26:221–233.e6. 10.1016/j.stem.2019.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gagne JP, Gagne P, Hunter JM, Bonicalzi ME, Lemay JF, Kelly I, Le Page C, Provencher D, Mes-Masson AM, Droit A, Bourgais D, Poirier GG. 2005. Proteome profiling of human epithelial ovarian cancer cell line TOV-112D. Mol Cell Biochem 275:25–55. 10.1007/s11010-005-7556-1. [DOI] [PubMed] [Google Scholar]
- 70.Kim NS, Hahn Y, Oh JH, Lee JY, Oh KJ, Kim JM, Park HS, Kim S, Song KS, Rho SM, Yoo HS, Kim YS. 2004. Gene cataloging and expression profiling in human gastric cancer cells by expressed sequence tags. Genomics 83:1024–1045. 10.1016/j.ygeno.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 71.Robinson WS. 1992. The role of hepatitis B virus in the development of primary hepatocellular carcinoma. Part I. J Gastroenterol Hepatol 7:622–638. 10.1111/j.1440-1746.1992.tb01497.x. [DOI] [PubMed] [Google Scholar]
- 72.Wadle A, Mischo A, Henrich PP, Stenner-Lieven F, Scherer C, Imig J, Petersen G, Pfreundschuh M, Renner C. 2005. Characterization of Hap/BAG-1 variants as RP1 binding proteins with antiapoptotic activity. Int J Cancer 117:896–904. 10.1002/ijc.21259. [DOI] [PubMed] [Google Scholar]
- 73.Yang WL, Wei L, Huang WQ, Li R, Shen WY, Liu JY, Xu JM, Li B, Qin Y. 2014. Vigilin is overexpressed in hepatocellular carcinoma and is required for HCC cell proliferation and tumor growth. Oncol Rep 31:2328–2334. 10.3892/or.2014.3111. [DOI] [PubMed] [Google Scholar]
- 74.Kinzler KW, Vogelstein B. 1997. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature 386:761–763. 10.1038/386761a0. [DOI] [PubMed] [Google Scholar]
- 75.Bacolla A, Sengupta S, Ye Z, Yang C, Mitra J, De-Paula RB, Hegde ML, Ahmed Z, Mort M, Cooper DN, Mitra S, Tainer JA. 2021. Heritable pattern of oxidized DNA base repair coincides with pre-targeting of repair complexes to open chromatin. Nucleic Acids Res 49:221–243. 10.1093/nar/gkaa1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dutertre M, Vagner S. 2017. DNA-damage response RNA-binding proteins (DDRBPs): perspectives from a new class of proteins and their RNA targets. J Mol Biol 429:3139–3145. 10.1016/j.jmb.2016.09.019. [DOI] [PubMed] [Google Scholar]
- 77.Dutertre M, Lambert S, Carreira A, Amor-Gueret M, Vagner S. 2014. DNA damage: RNA-binding proteins protect from near and far. Trends Biochem Sci 39:141–149. 10.1016/j.tibs.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 78.Scott DD, Trahan C, Zindy PJ, Aguilar LC, Delubac MY, Van Nostrand EL, Adivarahan S, Wei KE, Yeo GW, Zenklusen D, Oeffinger M. 2017. Nol12 is a multifunctional RNA binding protein at the nexus of RNA and DNA metabolism. Nucleic Acids Res 45:12509–12528. 10.1093/nar/gkx963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beli P, Lukashchuk N, Wagner SA, Weinert BT, Olsen JV, Baskcomb L, Mann M, Jackson SP, Choudhary C. 2012. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol Cell 46:212–225. 10.1016/j.molcel.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ. 2012. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol 14:318–328. 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mattoo AR, Pandita RK, Chakraborty S, Charaka V, Mujoo K, Hunt CR, Pandita TK. 2017. MCL-1 depletion impairs DNA double-strand break repair and reinitiation of stalled DNA replication forks. Mol Cell Biol 37:e00535-16. 10.1128/MCB.00535-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pandita TK, Pathak S, Geard CR. 1995. Chromosome end associations, telomeres and telomerase activity in ataxia telangiectasia cells. Cytogenet Cell Genet 71:86–93. 10.1159/000134069. [DOI] [PubMed] [Google Scholar]
- 83.Gupta A, Hunt CR, Hegde ML, Chakraborty S, Udayakumar D, Horikoshi N, Singh M, Ramnarain DB, Hittelman WN, Namjoshi S, Asaithamby A, Hazra TK, Ludwig T, Pandita RK, Tyler JK, Pandita TK. 2014. MOF phosphorylation by ATM regulates 53BP1-mediated double-strand break repair pathway choice. Cell Rep 8:177–189. 10.1016/j.celrep.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Altaf M, Utley RT, Lacoste N, Tan S, Briggs SD, Cote J. 2007. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol Cell 28:1002–1014. 10.1016/j.molcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Torres MJ, Pandita RK, Kulak O, Kumar R, Formstecher E, Horikoshi N, Mujoo K, Hunt CR, Zhao Y, Lum L, Zaman A, Yeaman C, White MA, Pandita TK. 2015. Role of the exocyst complex component Sec6/8 in genomic stability. Mol Cell Biol 35:3633–3645. 10.1128/MCB.00768-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study includes no data deposited in external repositories.