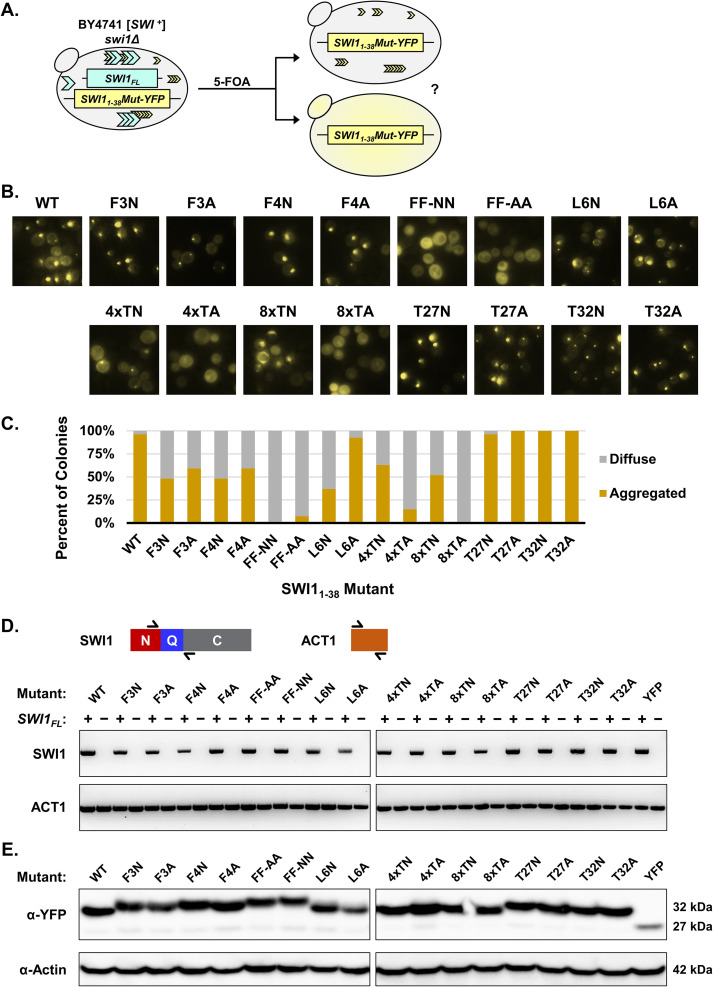
FIG 3.
Mutation of both phenylalanine residues leads to Swi11–38 being unable to maintain the prion fold in the absence of Swi1FL. (A) Diagram of the experiment. BY4741 swi1Δ/p416TEF-SWI1FL/p415TEF-SWI11–38Mut-YFP [SWI+] cells containing aggregates were treated with 5-FOA to select against cells containing the p416TEF-SWI1FL plasmid. The resulting BY4741 swi1Δ/p415TEF-SWI11–38Mut-YFP cells were observed using fluorescence microscopy for aggregate foci or diffuse signals. (B) Representative fluorescence images of the resulting BY4741 swi1Δ/p415TEF-SWI11–38Mut-YFP cells. See Table 1 for amino acid sequences of the mutants. For each construct, 3 aggregate-containing BY4741 swi1Δ/p416TEF-SWI1FL/p415TEF-SWI11–38Mut-YFP isolates were treated with 5-FOA to drop out the full-length Swi1 expression plasmid. From there, 9 colonies for each isolate (for a total of 27 colonies) were examined for each construct. Shown are representative views, and quantitative results are shown in panel C. (C) Quantification of yeast colonies retaining Swi11–38Mut-YFP aggregates after the removal of Swi1FL via 5-FOA treatment. For experiments quantified in this graph, aggregated colonies had >25% of cells containing aggregates, and diffuse colonies showed aggregation in <5% of cells. The percentage of colonies was calculated as the number of each of the two types of colonies (aggregated and diffuse) divided by the total number of colonies examined for each construct. (D) Diagram of RT-PCR primer targets and the resulting RT-PCR amplification visualized by agarose gel. Primers flanking the Q region of SWI1 were used to confirm the loss of SWI1FL in the BY4741 swi1Δ/p415TEF-SWI11–38Mut-YFP cells. Primers covering ACT1 were used as a positive control. Samples labeled as + correspond to the pre-5-FOA BY4741 swi1Δ/p415TEF-SWI11–38Mut-YFP cells. Samples labeled as − correspond to the post-5-FOA BY4741 swi1Δ/p415TEF-SWI11–38Mut-YFP cells. (E) Western blot of BY4741 swi1Δ/p415TEF-SWI11–38Mut-YFP cells. The membrane was probed with either anti-GFP or antiactin. Estimated molecular weights based on the sequence are listed at the right.