Fig. 3.
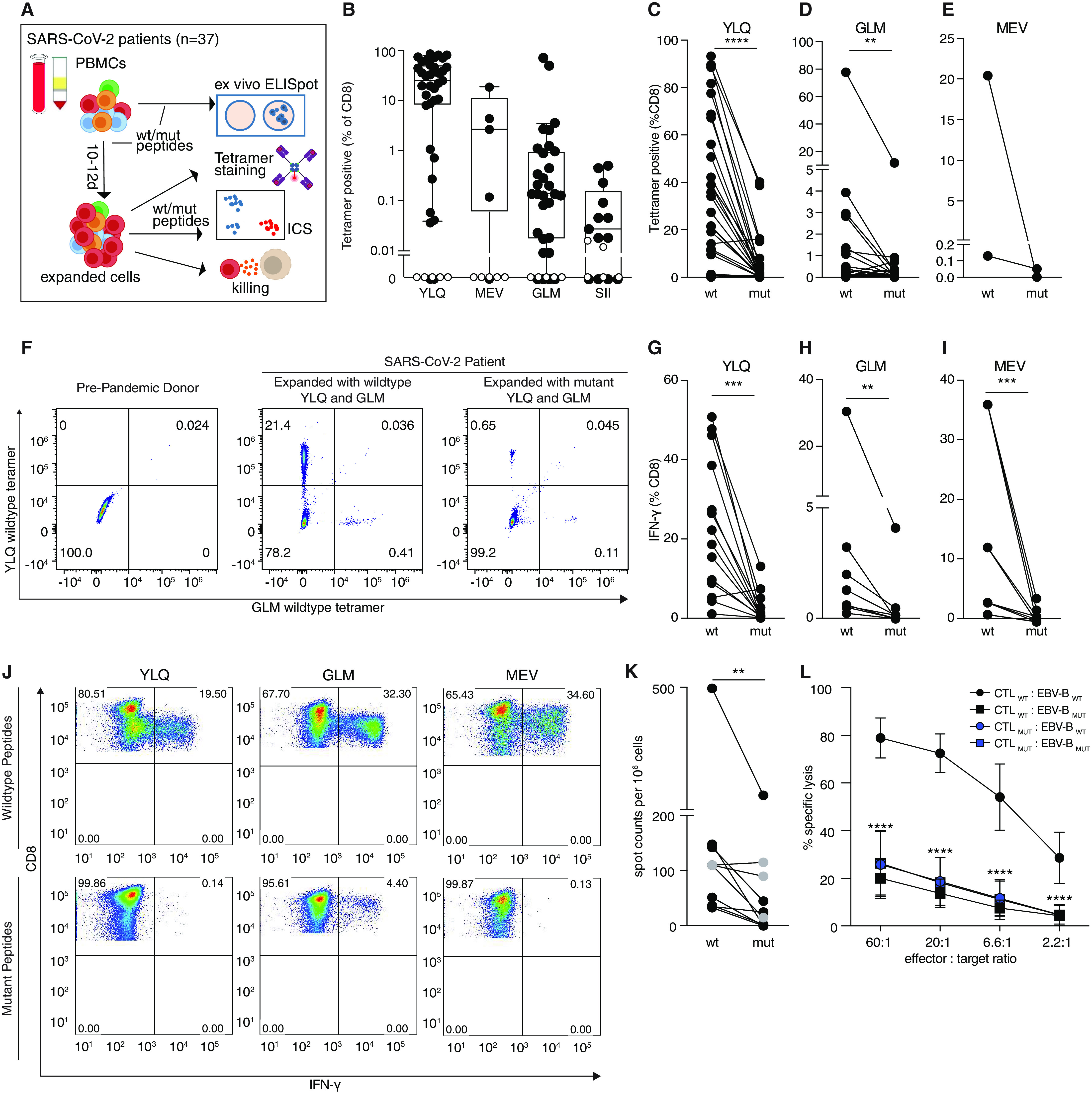
SARS-CoV-2 epitope mutations are associated with decreased CTL responses. A) Experimental overview. B) CTL responses against wild type epitopes. PBMCs were isolated from HLA-A*02:01 or HLA-B*40:01 positive SARS-CoV-2 patients (black, n=35, 5, 3, or 13 respectively, or pre-pandemic controls with unknown HLA status (white, n=7), expanded 10-12 days with indicated peptides, and stained with wild type tetramers. Boxes show median ± 25th and 75th percentile and whiskers indicate 10th and 90th percentile. C-E) T cells expanded with mutant peptides do not give rise to wild type peptide-specific CTLs. PBMCs were isolated as in B), stimulated with wild type or mutant peptides and stained with tetramers containing the wild type peptide. (n=27, 25, and 2 patients per epitope). F) Representative FACS plots for C-E. G-I) Impact of mutations on CTL response. PBMCs expanded with wild type or mutant peptides as indicated, were analyzed for IFN-γ-production via ICS after restimulation with wild type or mutant peptide (n=14, 8, and 4 patients per epitope). J) Representative FACS plots for G-I. K) Ex vivo IFN-γ ELISpot assays from PBMCs stimulated with the YLQ peptide or the corresponding mutant (n=7, PBMCs obtained 2.7 ± 0.8 weeks after symptom onset) or the MEV peptide (marked in gray) or corresponding mutant (n=1, PBMCs obtained 3 weeks after symptom onset). Two or three wells were evaluated per sample and peptide. Patient ID is as indicated in Table S6. L) CTL killing assay. PBMCs from 4 patients were expanded with wild type or mutant YLQ peptide, mixed with autologous EBV+ B cells that were pulsed with wild type or mutant YLQ peptide and specific killing was assessed (n=2 per patient). Error bars represent mean ± SD. Significance is indicated as *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, tested by Wilcoxon matched-pairs signed rank test (C,D,E,G,H,I,K) or 2-way ANOVA followed by Dunnett’s multiple comparison test (L).
