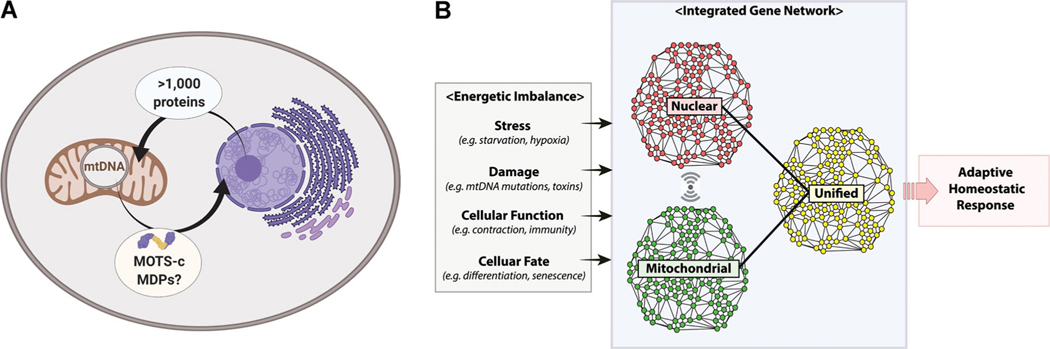
Figure 1.
Mitonuclear genome cross-regulation and the composite gene network. A) The mitochondrial proteome consists of over 1000 proteins that are encoded in the nuclear genome. On the contrary, the nuclear proteome was devoid of regulatory factors encoded in the mitochondrial genome. MOTS-c is the first MDP that has been shown to enter the nucleus and directly engage in adaptive nuclear gene expression. MOTS-c may be a harbinger of many other MDPs with nuclear roles. This notion would be consistent with the observation that the coevolved mitonuclear genomes independently encode for reciprocal gene-regulatory factors to coordinate gene regulation. B) In line with (A), eukaryotic gene networks are dynamically built based on a unified, yet split, bipartite genome compartmentalized in the nucleus and mitochondria. The unified mitonuclear gene network may provide a richer cellular context during adaptive gene responses to diverse pathophysiological cellular stimuli (stress, damage, function, fate) that are often accompanied by energetic imbalance. Thus, integrated gene analyses that account for the interaction of the two genomes would provide a more comprehensive biological perspective.