Abstract
Chemical investigation of the South China Sea soft coral Lemnalia sp. afforded 13 structurally diverse terpenoids, including three new neolemnane sesquiterpene lineolemnenes E–G (1–3); a new aristolane-type sesquiterpenoid, 2-acetoxy-aristolane (4); four new decalin-type bicyclic diterpenes, named biofloranates A−D (5−8); a new serrulatane, named euplexaurene D (9); and a new aromadendrane-type diterpenoid cneorubin K (10), together with three known related compounds (11−13). The structures of the new compounds were elucidated by NMR spectroscopy, the Mosher’s method, and ECD analysis. Compounds 1–13 were tested in a wide panel of biological assays. Lineolemnene J (3) showed weak cytotoxicity against the CCRF-CEM cancer cell line. The isolated new diterpenes, except euplexaurene D (9), demonstrated moderate antimicrobial activity against Bacillus subtilis and Staphylococcus aureus with a MIC of 4−64 μg/mL. Compound 2 exhibited a mild inhibitory effect against influenza A H1N1 virus (IC50 = 5.9 µM).
Keywords: soft coral, Lemnalia sp., terpenoids, antimicrobial activity, cytotoxicity
1. Introduction
In the competition for food and living space for survival, marine organisms accumulate and secrete secondary metabolites that are deterrent, offensive, or even toxic to other organisms—that is, chemical defense substances [1]. Marine invertebrates, such as soft corals, grow slowly and lack physical defense capabilities, but they can survive in competitive marine environments. What they often depend on are chemical defense strategies, which represent a rich source of unexplored chemical structures and biological diversity with great potential for drug discovery [2].
The South China Sea is located in the tropics and subtropics, with a vast sea area and rich marine biological resources. It is a sea area with numerous coral distributions. Since the 1980s, researchers have studied the chemical composition of many kinds of corals collected from the South China Sea [3,4,5,6,7,8]. Many compounds with novel structures have been isolated, including steroids, terpenoids, alkaloids, etc. Among them, the marine soft coral genus Lemnalia has afforded structurally diverse novel terpenoids, including sesquiterpenes and diterpene glycosides, with promising biological activity [7].
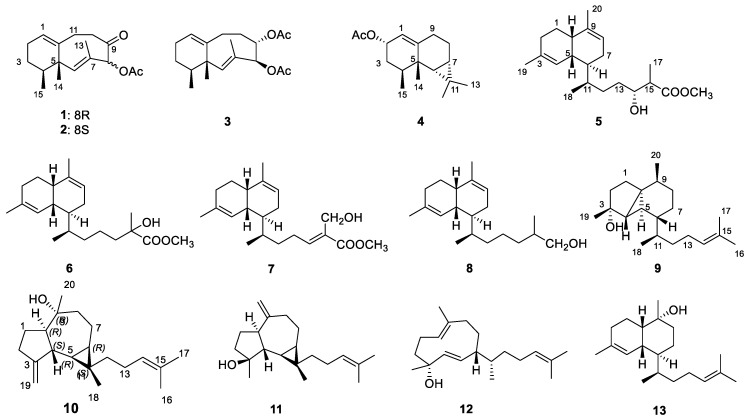
In the course of our efforts to search for novel and bioactive natural marine products from soft corals and find more diverse and complex terpenoids, we obtained Lemnalia sp. (No. XSSC201907), a soft coral collected from the coast of Xisha Islands in the South China Sea. The acetone extracts were chemically investigated, resulting in the discovery of 13 structurally diverse terpenoids, including four new sesquiterpenes (1–4), six new diterpenoids (5–10), and three known related compounds (11–13) (Figure 1). In vitro cytotoxic, antimicrobial, and antiviral activity of the isolated compounds were investigated. Herein, their isolation, structure, and biological activity are described.
Figure 1.
Structures of compounds 1–13.
2. Results and Discussion
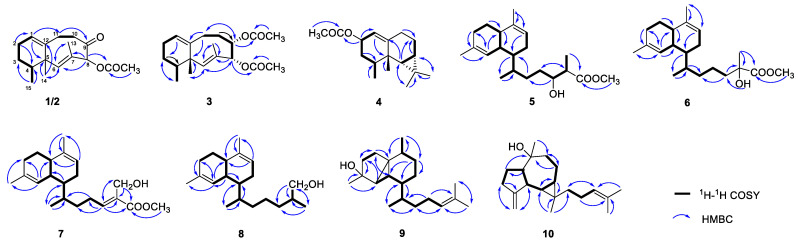
Compound 1 was isolated as a colorless oil; its HR-ESI-MS exhibited an ion peak at m/z 299.1616 [M + Na]+, corresponding to the molecular formula of C17H24O3Na (six degrees of unsaturation). The 13C NMR spectrum of 1 showed 17 signals (Table 1), in combined 1H NMR and HSQC experiments, indicating the presence of a ketone group (δC 201.2), an acetyl group (δC 170.1 and δC/δH 20.8/2.18), two double bonds (δC 140.4 and δC/δH 123.4/5.43; δC 127.8 and δC/δH 138.6/5.82), an oxygenated methine (δC/δH 85.5/5.66), a nonprotonated carbon (δC 46.2), two methylenes, four methines, and four methyls. The above signals revealed that 1 was a bicyclic structure. The neolemnane skeleton was established by extensive 2D NMR analysis [9]. The 1H-1H COSY spectrum revealed the connectivity of H-1/H2-2/H2-3/H-4/H3-15 and H2-10/H2-11 (Figure 2). The vinyl methyl (δC/δH 13.0/1.51) attached to C-7 (δC 127.8) was confirmed by HMBC correlations from H3-13 to C-6, C-7, and C-8. Together with further HMBC correlations from H-8 to C-6/C-7/C-13 and the carbonyl of the ester group, the planar structure of 1 was established. Moreover, established by HR-ESI-MS, compound 2 was found to possess the same molecular formula as 1. A comparison of the 1D and 2D NMR spectral data of 2 with those of 1 disclosed that they had the same planar structure.
Table 1.
NMR spectroscopic data (1H 600 MHz, 13C 150 MHz, CDCl3) for compounds 1–4.
| 1 | 2 | 3 | 4 | |||||
|---|---|---|---|---|---|---|---|---|
| Position | δH, Mult. | δC, Type | δH, Mult. | δC, Type | δH, Mult. | δC, Type | δH, Mult. | δC, Type |
| 1 | 5.43, m | 123.4, CH | 5.49, m | 125.1, CH | 5.46, m | 123.9, CH | 5.17, br.s | 118.1, CH |
| 2a | 2.07, m | 26.1, CH2 | 2.09, m | 26.0, CH2 | 2.06, m | 26.0, CH2 | 5.35, m | 68.9, CH |
| 2b | 2.01, m | — | 2.09, m | — | 2.06, m | — | — | — |
| 3a | 1.49, m | 26.6, CH2 | 1.51, m | 26.1, CH2 | 1.48, m | 26.7, CH2 | 1.93, m | 42.6, CH2 |
| 3b | 1.37, m | — | 1.51, m | — | 1.41, m | — | 1.57, m | — |
| 4 | 2.26, ov | 36.0, CH | 2.41, m | 33.2, CH | 2.28, m | 35.3, CH | 1.95, m | 37.9, CH |
| 5 | — | 46.2, C | — | 45.3, C | — | 43.6, C | — | 51.7, C |
| 6 | 5.82, s | 138.6, CH | 5.22, s | 147.5, CH | 5.33, s | 132.4, CH | 0.85, d (6.5) | 38.8, CH |
| 7 | — | 127.8, C | — | 128.0, C | — | 137.4, C | 1.22, br.t (6.3) | 32.4, CH |
| 8a | 5.66, s | 85.5, CH | 5.16, s | 84.8, CH | 5.67, d (6.9) | 73.7, CH | 2.01, m | 26.8, CH2 |
| 8b | — | — | — | — | — | — | 1.74, m | — |
| 9a | — | 201.2, C | — | 203.2, C | 4.86, dd (9.9, 7.3) | 79.5, CH | 1.90, m | 33.0, CH2 |
| 9b | — | — | — | — | — | — | 1.59, m | — |
| 10a | 2.75, td (12.6, 3.8) | 40.8, CH2 | 2.38, m | 42.9, CH2 | 1.74, m | 32.2, CH2 | — | 145.9, C |
| 10b | 2.16, m | — | 2.08, m | — | 1.57, dd (15.1, 9.3) | — | — | — |
| 11a | 2.59, m | 32.6, CH2 | 2.33, m | 34.3, CH2 | 2.22, dd (15.2, 9.4) | 34.6, CH2 | — | 20.3, C |
| 11b | 2.30, ov | — | 2.32, m | — | 2.05, m | — | — | — |
| 12 | — | 140.4, C | — | 142.4, C | — | 143.8, C | 1.00, s | 28.8, CH3 |
| 13 | 1.51, s | 13.0, CH3 | 2.01, s | 21.2, CH3 | 1.72, s | 15.5, CH3 | 1.14, s | 17.1, CH3 |
| 14 | 1.13, s | 21.4, CH3 | 1.03, s | 21.0, CH3 | 1.05, s | 22.1, CH3 | 1.76, s | 20.4, CH3 |
| 15 | 0.99, d (6.9) | 17.8, CH3 | 0.95, d (6.9) | 17.5, CH3 | 0.91, d (6.9) | 17.6, CH3 | 1.07, d (7.1) | 17.0, CH3 |
| 7-OAc | — | — | — | — | — | — | — | 171.1, C |
| — | — | — | — | — | — | 2.04, s | 21.5, CH3 | |
| 8-OAc | — | 170.1, C | — | 170.6, C | — | 170.1, C | — | — |
| 2.18, s | 20.8, CH3 | 2.23, s | 20.9, CH3 | 2.01, s | 20.9, CH3 | — | — | |
| 9-OAc | — | — | — | — | — | 169.8, C | — | — |
| — | — | — | — | 2.12, s | 20.9, CH3 | — | — | |
ov Overlapped signals, — absence of value.
Figure 2.
Key 1H-1H COSY and HMBC correlations of 1–10.
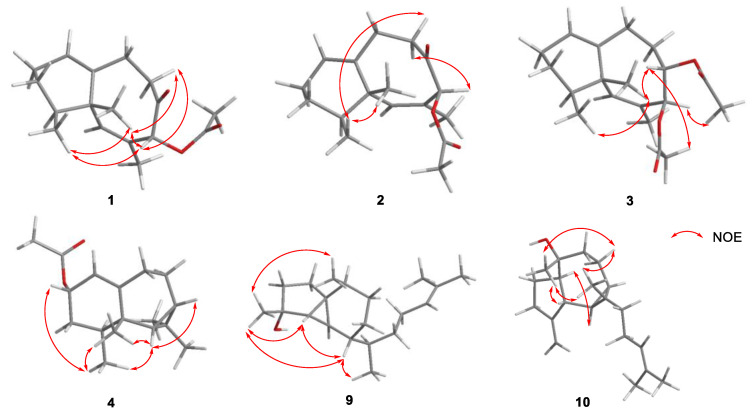
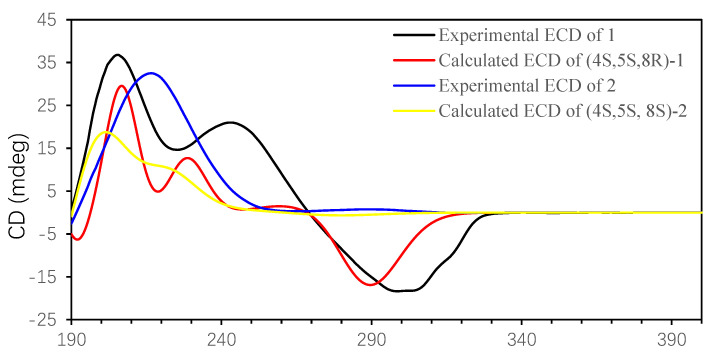
The geometry of the 6,7-double bonds of compounds 1 and 2 was elucidated from NOE correlations (Figure 3). The absence of an NOE correlation for H3-13/H-6 suggests an E geometry of the 6,7-double bond, which was different from the known lineolemnene A and B [10]. For compound 1, the NOESY correlations from H3-14 to H3-15, H-10a (δH 2.74) and from H-8 to H3-14 and H-10a (δH 2.75) placed H-8, H3-14, and H3-15 on the same face. Thus, the relative configuration of 1 was elucidated as 4R*,5R*,8S*. For compound 2, H3-15 was found to show NOE correlations with H3-14 and H-10a (δH 2.38), and H-8 showed an NOE correlation with H-10b (δH 2.08) but not with H3-14, suggesting that the relative configuration of 2 was 4S*,5S*,8S*. The absolute configurations (AC) of compounds 1 and 2 could be deduced from TDDFT/ECD calculations. A comparison of the experimental ECD spectrum of 1, 2, calculated (4S,5S,8R)-1 and calculated (4S,5S,8S)-2 (Figure 4) revealed that the AC of 1 and 2 was 4S,5S,8R and 4S,5S,8S, respectively. The NMR calculations were also conducted at the GIAO/mPW1PW91/6-31G (d, p) level. Finally, computed and experimental data were correlated, and the corresponding DP4+ probabilities were estimated. As shown in Figures S1 and S2, the experimental NMR data of compound 1 gave the best match for (4S,5S,8R)-1, and compound 2 gave the best match for (4S,5S,8S)-2.
Figure 3.
Key NOESY correlations of 1–4 and 9–10.
Figure 4.
Experimental and calculated ECD spectra of compounds 1 and 2.
The molecular formula of 3, C19H28O4, was established by HR-ESI-MS, requiring six degrees of unsaturation. The NMR data indicated the presence of two acetyl groups (δC/δH 170.1 and 20.9/2.01; δC/δH 169.8 and 20.9/2.12), and it was analogous of neolemnane [9]. Two oxygenated methines (δC/δH 79.5/4.86; δC/δH 73.7/5.67) were found according to the 1D NMR spectra. The placement of two acetyl groups at C-8 and C-9 was confirmed by HMBC correlations from H-8 (δH 5.67) to an acetyl carbon (δC 170.1), and from H-9 (δH 4.86) to another acetyl carbon (δC 169.8). The structure was further confirmed with the assistance of 1H-1H COSY and HMBC spectra, as shown in Figure 2. The E geometry of 6,7-double bond was deduced in the same manner as compounds 1 and 2. The relative stereochemistry of 3 was also deduced by the key NOE interactions. The observed NOE correlation (Figure 3) between H3-14/H3-15, H3-14/H-9, H-9/8-OAc, and H-8/9-OAc and no NOE correlation with H3-14/H-8 demonstrated that 8-OAc, H-9, H3-14, and H3-15 were close in space while H-8 and 9-OAc were distant, with an antiperiplanar orientation. As a result, the AC of 3 was proposed to be 4S,5S,8S,9S according to the NOE correlations and considering biosynthetic properties [9]. Compound 3 was the C-8 and C-9 epimer of lineolemnene C [10].
Compound 4 was also obtained as a colorless oil. The molecular formula was established as C17H26O2 by HR-ESI-MS (m/z 285.1824 [M + Na]+). The NMR signals (δC/δH 118.1/5.17 and 145.9) (Table 1) showed the presence of a trisubstituted double bond. The 1H–1H COSY spectrum revealed the connectivity of H-1/H-2/H2-3/H-4/H3-15 and H-6/H-7/H2-8/H2-9, indicating that it was an aristolane-type sesquiterpenoid [11]. The location of an acetyl group (δC/δH 171.1 and 21.5/2.04) at position C-2 (δC/δH 68.9/5.35) was revealed by the relevant HMBC correlations. The third ring was a cyclopropane due to the key H3-12/C-6/C-7/C-11 and H3-13/C-6/C-7/C-11 HMBC correlations. The NOE correlations between H-2/H3-15, H3-15/H3-14, and H-6/H3-14 revealed the relative configuration of 4. ECD calculation was performed (Figure S3) to determine the absolute configuration of compound 4 as 2S,4S,5S,6R,7S. Compound 4 was named 2-acetoxy-aristolane. The acetoxy group was of natural origin but not artifact, which was confirmed by LC-HRMS analysis of the methanol extract of Lemnalia sp. (Figure S37). The protonated- molecular ion peak[M + H]+ of 4 was extracted successfully (Figure S37).
Biofloranate A (5) was also a colorless oil, and the molecular formula was established as C21H34O3 (HR-ESI-MS: m/z 357.2400 [M + Na]+). The 1D NMR and HSQC spectra of 5 (Table 2 and Table 3) disclosed two trisubstituted olefinic bonds (δC/δH 134.7 and 123.6/5.47; δC/δH 136.7 and 121.2/5.39), an acetate group (δC/δH 176.7 and 51.7/3.71), and an oxygenated methine (δC/δH 72.1/3.85), which also indicated a bicyclic structure. A comparison of the 1H and 13C NMR data of 5 with the aglycone of lemnabourside [12] showed that they had identical ring systems, including stereochemistry, and similar side chains. The planar structure was further established by the analysis of COSY and HMBC correlations. The orientation of 14-OH of 5 was assigned via Mosher’s method. Esterification of 5 with (R)- and (S)-MTPA chloride occurred at the C-14 hydroxyl group to give (S)- and (R)-MTPA ester derivatives. The observed ΔδH (S-R) value distribution pattern (Figure 5, Figures S82 and S83) established the 14R absolute configuration for 5. However, the configuration of C-15 was difficult to determine.
Table 2.
1H NMR (600 MHz) spectroscopic data for compounds 5–10.
| Position | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|
| δH, Mult. | δH, Mult. | δH, Mult. | δH, Mult. | δH, Mult. | δH, Mult. | |
| 1a | 1.85, m | 1.84, m | 1.78, m | 1.84, ov | 1.84, m | 1.88, ov |
| 1b | 1.36, ov | 1.36, m | 1.36, ov | 1.37, ov | 1.52, ov | 1.47, qd (11.5, 8.3) |
| 2a | 1.98, ov | 1.97, m | 1.99, ov | 1.98, ov | 1.53, ov | 2.40, m |
| 2b | 1.92, ov | 1.91, ov | 1.95, ov | 1.91, ov | 1.35, ov | 2.31, m |
| 4 | 5.47, br.s | 5.47, br.s | 5.45, br.s | 5.48, br.s | 0.87, ov | 1.71, ov |
| 5 | 2.04, dd (9.3, 4.7) | 2.02, m | 2.04, ov | 2.04, m | 0.80, ov | 0.54, br.t (9.5) |
| 6 | 1.52, ov | 1.48, m | 1.51, dt (7.4, 3.1) | 1.51, m | 1.14, m | 0.66, td (10.9, 10.1, 6.5) |
| 7a | 1.78, ov | 1.75, ov | 1.83, ov | 1.84, ov | 1.33, ov | 1.85, ov |
| 7b | 1.78, ov | 1.75, ov | 1.78, ov | 1.77, ov | 0.83, ov | 0.89, m |
| 8a | 5.39, br.s | 5.39, br.s | 5.39, br.s | 5.40, br.s | 1.57, ov | 1.75, dd (12.9, 7.1) |
| 8b | — | — | — | — | 0.51, m | 1.60, ov |
| 9 | — | — | — | — | 1.64, dd (11.9, 6.0) | — |
| 10 | 1.95, ov | 1.93, ov | 1.94, ov | 1.94, ov | — | 1.92, td (11.1, 6.5) |
| 11 | 1.78, ov | 1.78, ov | 1.83, ov | 1.79, ov | 1.51, ov | — |
| 12a | 1.38, ov | 1.18, ov | 1.36, ov | 1.30, ov | 1.46, m | 1.26, m |
| 12b | 1.22, ov | 1.18, ov | 1.36, ov | 1.21, ov | 1.21, m | 1.16, m |
| 13a | 1.51, m | 1.69, ov | 2.27, m | 1.19, m | 2.04, dd (13.9, 6.3) | 2.06, m |
| 13b | 1.36, ov | 1.62, ov | 2.27, m | 1.19, m | 1.92, dq (14.5, 7.5) | 2.06, m |
| 14a | 3.85, dd (8.0, 3.8) | 1.39, ov | 6.88, t (7.8) | 1.37, ov | 5.12, br.t (6.2) | 5.09, t (7.1) |
| 14b | — | 1.15, m | — | 1.08, ov | — | — |
| 15 | 2.54, qd (7.2, 3.5) | — | — | 1.61, m | — | — |
| 16a | — | — | — | 3.40, dd (10.4, 6.6) | 1.61, s | 1.60, s |
| 16b | — | — | — | 3.49, dd (10.5, 5.9) | — | — |
| 17 | 1.18, d (7.2) | 1.39, s | 4.33, br.s | 0.90, d (6.7) | 1.69, s | 1.67, s |
| 18 | 0.83, d (6.8) | 0.79, d (6.7) | 0.84, d (6.9) | 0.81, d (6.8) | 0.90, d (6.7) | 1.04, s |
| 19a | 1.68, s | 1.69, s | 1.70, s | 1.69, s | 1.28, s | 4.90, s |
| 19b | — | — | — | — | — | 4,79, s |
| 20 | 1.68, s | 1.68, s | 1.69, s | 1.69, s | 0.93, d (6.4) | 1.12, s |
| OMe | 3.71, s | 3.78, s | 3.78, s | — | — | — |
ov Overlapped signals, — absence of value.
Table 3.
13C NMR (150 MHz) spectroscopic data for compounds 5–10.
| Position | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|
| δC, Type | δC, Type | δC, Type | δC, Type | δC, Type | δC, Type | |
| 1 | 24.6, CH2 | 24.6, CH2 | 24.5, CH2 | 24.7, CH2 | 29.5, CH2 | 26.3, CH2 |
| 2 | 30.8, CH2 | 30.8, CH2 | 30.7, CH2 | 30.8, CH2 | 36.3, CH2 | 32.1, CH2 |
| 3 | 134.7, C | 134.6, C | 134.8, C | 134.5, C | 80.4, C | 157.8, C |
| 4 | 123.6, CH | 123.8, CH | 123.4, CH | 123.8, CH | 39.0, CH | 41.6, CH |
| 5 | 36.3, CH | 36.4, CH | 36.2, CH | 36.3, CH | 23.1, CH | 32.7, CH |
| 6 | 39.1, CH | 38.9, CH | 38.7, CH | 39.0, CH | 42.5, CH | 26.6, CH |
| 7 | 24.6, CH2 | 24.6, CH2 | 24.5, CH2 | 24.6, CH2 | 25.3, CH2 | 20.4, CH2 |
| 8 | 121.2, CH | 121.4, CH | 121.0, CH | 121.4, CH | 31.7, CH2 | 44.6, CH2 |
| 9 | 136.7, C | 136.7, C | 136.6, C | 136.6, C | 30.9, CH | 75.1, C |
| 10 | 39.5, CH | 39.6, CH | 39.4, CH | 39.5, CH | 33.6, C | 57.9, CH |
| 11 | 31.8, CH | 35.6, CH | 31.6, CH | 31.7, CH | 38.3, CH | 23.2, C |
| 12 | 31.4, CH2 | 31.6, CH2 | 34.5, CH2 | 35.9, CH2 | 34.6, CH2 | 43.2, CH2 |
| 13 | 31.9, CH2 | 21.7, CH2 | 26.6, CH2 | 25.0, CH2 | 26.1, CH2 | 25.4, CH2 |
| 14 | 72.1, CH | 40.6, CH2 | 146.1, CH | 33.5, CH2 | 125.0, CH | 124.8, CH |
| 15 | 44.0, CH | 74.8, C | 130.4, C | 35.8, CH | 131.3, C | 131.1, C |
| 16 | 176.7, C | 177.9, C | 168.0, C | 68.4, CH2 | 17.7, CH3 | 17.6, CH3 |
| 17 | 10.6, CH3 | 26.0, CH3 | 57.2, CH2 | 16.6, CH3 | 25.7, CH3 | 25.7, CH3 |
| 18 | 13.2, CH3 | 13.3, CH3 | 13.1, CH3 | 13.3, CH3 | 16.1, CH3 | 13.7, CH3 |
| 19 | 23.9, CH3 | 24.0, CH3 | 23.8, CH3 | 24.0, CH3 | 27.9, CH3 | 105.5, CH2 |
| 20 | 21.7, CH3 | 21.8, CH3 | 21.6, CH3 | 21.7, CH3 | 18.8, CH3 | 20.5, CH3 |
| OMe | 51.7, CH3 | 52.7, CH3 | 51.7, CH3 |
Figure 5.
ΔδH values (Δδ (in ppm) = δS-δR) obtained for (S)- and (R)-MTPA esters of compound 5 in pyridine-d5.
Compound 6 shared the same molecular formula as 5. The 1H and 13C NMR spectra of 6 (Table 2 and Table 3) were also similar to those of 5, except that the oxygenated carbon was quaternary (δC 74.8) instead of a methine. As a result, the hydroxy was connected to C-15 instead of C-14. The orientation of 15-OH present in 3 was hard to determine, leading to the unknown relative configuration of C-15. Compound 6 was given the name biofloranate B.
Biofloranate C (7) had a molecular formula of C21H32O3 as determined by HR-ESI-MS data, requiring six degrees of unsaturation. Comparing the molecular formula of 7 with 5 and 6, two hydrogens were missing, corresponding to the presence of one more trisubstituted double bond (δC/δH 130.4 and 146.1/6.88). An oxygenated methylene H-17 (δC/δH 57.2/4.33) showed HMBC correlations with C-14, C-15, and C-16. Key H2-13/H-14 1H-1H COSY correlation and H-14/C-12/C-13/C-15/C-16/C-17 HMBC correlations (Figure 2) suggested that the extra double bond was C-14−C-15. The H-14/16-OCH3 NOESY correlation placed H-14 and the methoxy group on the same side (Figure S59). Thus, the structure of 7 was established unambiguously. Biofloranate D (8) was obtained as a colorless oil and the formula was assigned as C20H34O by HR-ESI-MS (m/z 313.2336, [M + Na]+). The 1D NMR data of 8 (Table 2 and Table 3) also corresponded closely to those of aglycone of lemnabourside [12]. Compared with aglycone, the 1D NMR data revealed the absence of an aldehyde group but the presence of an oxygenated methylene (δC/δHa/δHb 68.4/3.49/3.40). The structure of 8 was also confirmed by 1H-1H COSY and HMBC correlations, as shown in Figure 2. The AC of C-15 was also difficult to determine.
Euplexaurene D (9) was isolated as a white powder, and HR-EI-MS analysis (found 290.2604, M+) showed the molecular formula C20H34O, suggesting four degrees of unsaturation. As the only sp2 group, the trisubstituted olefinic bond (δC/δH 131.3 and 125.0/5.12) was deduced from the 1D NMR data, indicating the tricyclic structure of 9. In the 13C NMR spectrum (Table 3), 20 carbon signals were assignable to three quaternary carbons, six methines, six methylenes, and five methyls. Comparing the 13C NMR spectrum of 9 with that of reported euplexaurene A [13] showed that they have identical ring systems, and the main difference was the position of the hydroxy group. Measuring the 2D NMR spectrum (Figure 2) enabled the assignment of all resonances in the 1H NMR and 13C NMR spectra (Table 2 and Table 3). The relative configurations of 9 were assigned by NOESY correlations between H3-19/H-4, H3-19/H3-20, H-6/H3-19, and H-6/H3-18 (Figure 4).
Cneorubin K (10) was obtained as a colorless oil, with the molecular formula C20H32O determined from HR-EI-MS (m/z 270.2334 [M − H2O]+), indicating five degrees of unsaturation. The NMR spectra revealed the presence of a trisubstituted olefinic bond (δC/δH 131.1 and 124.8/5.09) and an exocyclic double bond (δC/δH 157.8 and 105.5/4.90/4.79), which accounted for two degrees of unsaturation, and the remaining three degrees of unsaturation required a tricyclic system in 10. The aromadendrane diterpenoid skeleton [14] was established by extensive 1H-1H COSY and HMBC analysis, as shown in Figure 2. In particular, the exocyclic double bond was C-3/C-19 and oxygenated nonprotonated carbon was C-9 (δC 75.1), which were confirmed by HMBC correlations from H2-19 to C-2/C-3 and H3-20 to C-10/C-8. The relative stereochemistry of 10 was deduced by the key NOE interactions. The NOE correlations between H-5/H-10, H3-20/H-4, H3-20/H2-7b, H3-20/H3-18, H3-18/H2-7b and H3-18/H-4 revealed the relative configuration of 10 (Figure 3). ECD calculation was performed (Figure S4) to determine the absolute configuration of compound 10 as 4S,5R,6R,9R,10R,11S.
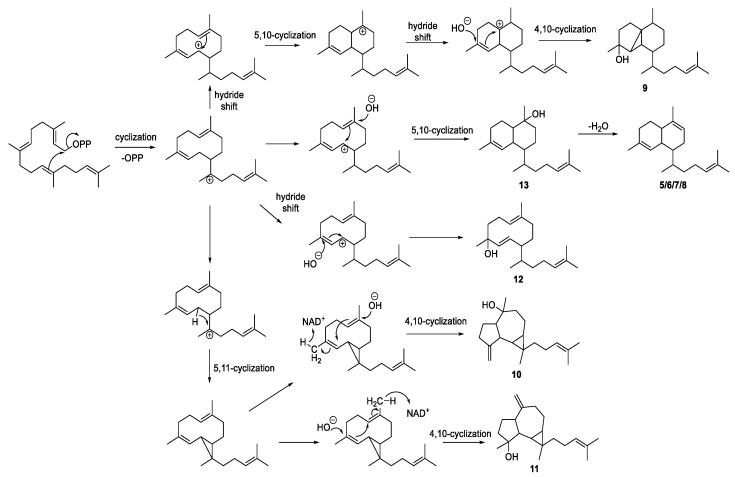
According to previously reports, all diterpenoids from Lemnalia have a biflorane-based skeleton [7]. In the present study, diterpenoids separated from the Lemnalia organism possessed different ring structures, characterized as biflorane, 5,3,6-tricyclic, 5,7,3-tricyclic, and 10-membered carbocyclic skeletons, and were biogenetically related to each other. A geranylgeranyl-PP (GGPP, C20) has been regarded as the same biosynthetic precursor of these diterpenoids, and we further propose biosynthesis for these diterpenoids, as illustrated in Scheme 1.
Scheme 1.
Plausible biosynthetic pathway of diterpenoids 5–13.
All of the isolated compounds (1–13) were evaluated by a wide panel of biological assays, including cytotoxic activity (A-549, HepG2, HeLa, and CCRF-CEM cell lines) and antibacterial (Bacillus subtilis and Staphylococcus aureus) and antiviral (H1N1 and HSV-1) activity (as shown in Table S1). As a result, lineolemnene J (3) showed weak cytotoxicity against the CCRF-CEM cancer cell line (IC50 15.9 µM). Most of the diterpenoids exhibited corresponding antibacterial activity against Bacillus subtilis and Staphylococcus aureus. Moreover, sesquiterpenes 1–4 exhibited anti-H1N1 virus activity with 77.6–100% inhibition at a concentration of 30 µM, and compound 2 exhibited activity with IC50 = 5.9 µM.
3. Materials and Methods
3.1. General Experimental Procedures
Optical rotations and UV spectra were measured on a Jasco P-1010 Polarimeter polarimeter (JASCO, Tokyo, Japan) and ThermoFisher Evolution 201/220 spectrophotometer (Thermo Scientific, Waltham, MA, USA). in MeOH at 20 °C., respectively. NMR spectra were recorded on a Bruker AVANCE NEO 600 spectrometer (BrukerBiospin AG, Fällanden, Germany) with a 5 mm inverse detection triple resonance (H-C/N/D) cryoprobe. 1H chemical shifts were referenced to the residual CDCl3 (7.24 ppm) and 13C chemical shifts were referenced to the CDCl3 (77.0 ppm) solvent peaks. High-resolution electrospray ionization mass spectra (HRESIMS) were performed on an Agilent 6230 TOF LC/MS system (Agilent Technologies Inc., Palo Alto, CA, USA) and Thermo Scientific TM Q Exactive PlusTM (Thermo Scientific, Waltham, MA, USA). High-resolution electron impact mass spectra (HREIMS) were recorded on JEOL AccuTOF LC-plus 4G (JEOL, Tokyo, Japan). Reversed-phase HPLC purifications were performed on a Shimadzu LC-20AT HPLC system equipped with a quaternary gradient solvent delivery unit and a photodiode array detector (Shimadzu, Kyoto, Japan), using a semipreparative Cosmosil ODS column (250 mm × 10.0 mm i.d., 5 µm, Cosmosil, Nakalai Tesque Co. Ltd., Kyoto, Japan). Column chromatography was performed on a silica gel (Qingdao Haiyang Chemical Co., Ltd., Qingdao, China) and Cosmosil 75 C18 (75 μm, Nakalai Tesque Co. Ltd., Kyoto, Japan).
3.2. Animal Material
Soft coral Lemnalia sp. (no. XSSC201907) was collected from the Xisha Islands of the South China Sea in April 2019 by scuba at a depth of 10 m. It was identified by Prof. Pingjyun Sung, National Museum of Marine Biology and Aquarium (NMMBA), Taiwan, China.
3.3. Extraction and Isolation
A sample of Lemnalia sp. (1.9 kg) was freeze-dried and extracted with acetone (1:1, v/v, 5 times). Then the extracting solution was concentrated to yield 33.8 g of residue. The residue was suspended in H2O (500 mL) and partitioned with Et2O (500 mL, 4 times) to yield an Et2O solvent extract (25.5 g). The Et2O solvent extract was separated on a Sephadex LH-20 column eluted with petroleum ether (PE)-CH2Cl2-MeOH (2:1:1, v/v/v), leading to 4 fractions (Fr.1–Fr.4). Fr.2 (10.4 g) was chromatographed over a silica gel column (PE/EtOAc, from 50:1 to 1:1) to give 9 fractions (Fr.2.1−Fr.2.9). Fr.2.3 was further separated on an ODS column (MeCN/H2O, from 50 to 100%) and purified by semipreparative HPLC (MeOH/H2O, 97:3, 3 mL/min) to give compound 8 (15.7 mg). Fr.2.5 was also separated on the ODS column (MeCN/H2O, from 40 to 100%) to give 4 fractions (Fr.2.4.1–Fr.2.4.4). Fr.2.4.2 was purified by semipreparative HPLC (MeOH/H2O, 87:13, 3 mL/min) to give compounds 9 (12.0 mg), 10 (4.1 mg), 11 (7.8 mg), and 13 (7.8 mg). Fr.2.4.3 was purified by semipreparative HPLC (MeOH/H2O, 92:8, 3 mL/min) to give compounds 6 (10.0 mg) and 12 (3.5 mg). Fr.2.6 was further separated on the ODS column (MeCN/H2O, from 40 to 100%) and purified by semipreparative HPLC (MeOH/H2O, 88:12, 3 mL/min) to give compounds 4 (9.8 mg), 5 (10.0 mg), and 7 (5.0 mg). Fr.2.8 was separated on the ODS column (MeCN/H2O, from 40 to 80%) and purified by semipreparative HPLC (MeOH/H2O, 75:25, 3 mL/min) to give compounds 1 (11.0 mg), 2 (23.5 mg), and 3 (28.0 mg).
Lineolemnene E (1): colorless oil; { −21 (c 0.1, MeOH)}; UV (MeOH) λmax (log ε) 204 (2.53) nm; 1H and 13C NMR spectroscopic data, Table 1; HRESIMS m/z 299.1616 [M + Na]+ (calcd for C17H24O3Na, 299.1623).
Lineolemnene F (2): colorless oil; { +214 (c 0.1, MeOH)}; UV (MeOH) λmax (log ε) 202 (2.49) nm; 1H and 13C NMR spectroscopic data, Table 1; HRESIMS m/z 299.1618 [M + Na]+ (calcd for C17H24O3Na, 299.1623).
Lineolemnene G (3): colorless oil; { +39 (c 0.1, MeOH)}; UV (MeOH) λmax (log ε) 202 (2.42) nm; 1H and 13C NMR spectroscopic data, Table 1; HRESIMS m/z 343.1880 [M + Na]+ (calcd for C19H28O4, 343.1886).
2-Aristolanol acetate (4): colorless oil; { −31 (c 0.1, MeOH)}; UV (MeOH) λmax (log ε) 203 (2.24) nm; 1H and 13C NMR spectroscopic data, Table 1; HRESIMS m/z 285.1824 [M + Na]+ (calcd for C17H26O2, 285.1831).
Biofloranate A (5): colorless oil; { +34 (c 0.1, MeOH)}; UV (MeOH) λmax (log ε) 202 (2.03) nm; 1H and 13C NMR spectroscopic data, Table 2 and Table 3; HRESIMS m/z 357.2400 [M + Na]+ (calcd for C21H34O3, 357.2406).
Biofloranate B (6): colorless oil; { +33 (c 0.1, MeOH)}; UV (MeOH) λmax (log ε) 201 (2.05) nm; 1H and 13C NMR spectroscopic data, Table 2 and Table 3; HRESIMS m/z 357.2395 [M + Na]+ (calcd for C21H34O3, 357.2406).
Biofloranate C (7): colorless oil; { −105 (c 0.1, MeOH)}; UV (MeOH) λmax (log ε) 202 (2.03) nm; 1H and 13C NMR spectroscopic data, Table 2 and Table 3; HRESIMS m/z 355.2251 [M + Na]+ (calcd for C21H32O3, 355.2249).
Biofloranate D (8): colorless oil; { +30 (c 0.1, MeOH)}; UV (MeOH) λmax (log ε) 202 (2.05) nm; 1H and 13C NMR spectroscopic data, Table 2 and Table 3; HRESIMS m/z 313.2511 (calcd for C20H34O, 313.2508).
Euplexaurene D (9): colorless oil; { +21 (c 0.1, MeOH)}; UV (MeOH) λmax (log ε) 201 (1.98) nm; 1H and 13C NMR spectroscopic data, Table 2 and Table 3; HREIMS m/z 290.2604 [M]+ (calcd for C20H34O, 290.2610).
Cneorubin K (10): colorless oil; { −24 (c 0.1, MeOH)}; UV (MeOH) λmax (log ε) 202 (2.04) nm; 1H and 13C NMR spectroscopic data, Table 2 and Table 3; HREIMS m/z 270.2334 [M-H2O]+ (calcd for C20H30, 270.2348).
3.4. Computational Methods
Geometry optimization was performed using Gaussian 09W (Gaussian Inc., Wallingford, CT, USA) and the B3LYP functional at the 6-311+G(d,p) level of theory as the method described in a reported article [15].
The theoretical calculation of 13C NMR chemical shifts of epimers 1 and 2 were carried out by geometry optimization [16]. The molecular structures were first minimized by MMFF. All torsional angles were systematically varied to search for the global minimum. The molecular mechanics minimized structures were then imported to Gaussian for all subsequent steps. Geometry optimization was conducted at the B3LYP/6-31G (d, p) level of theory. IEF-PCM solvation in chloroform was used for the stereoisomers. Chloroform was the solvent in which experimental NMR spectra were acquired. 13C NMR shielding constants were calculated at the GIAO/mPW1PW91/6-31G (d, p) level, with IEF-PCM solvation in the chloroform solvent.
3.5. Cytotoxic Activity Assay
Cell lines were purchased from the American Type Cultural Collection (ATCC, Manassas, VA, USA). The cytotoxicity of the compounds was evaluated against the A549, HepG2, Hela and CCRF-CEM with the Cell Counting Kit-8 (CCK-8) as the reported method [17]. Chidamide was used as a positive control.
3.6. Antibacterial Assays
The antibacterial activities were evaluated with the broth dilution assay [18]. Bacterial strains, Bacillus subtilis (CMCC (B) 63501), Staphylococcus aureus (CMCC (B) 26003) were used, and penicillin G as a positive control.
3.7. Antiviral Assays
The antiviral activities against the influenza A virus (H1N1) and (HSV-1) were evaluated as the literature reported [19]. Acyclovir was used as a positive control.
4. Conclusions
Marine invertebrates have been regarded as an important source of bioactive secondary metabolites. The acetone extracts of soft coral Lemnalia sp. (no. XSSC201907) were chemically investigated, resulting in the discovery of four new sesquiterpenes (1–4) and six new diterpenoids (5–10), along with three known related diterpenoids. Most of the isolated diterpenes were found to exhibit moderate antimicrobial activity against Bacillus subtilis and Staphylococcus aureus. Sesquiterpene lineolemnene J (3) showed weak cytotoxicity against the CCRF-CEM cancer cell line. Lineolemnene (2) exhibited medium inhibitory activity against influenza A H1N1 virus.
Acknowledgments
We thank Zhenhua Long, Daning Li, and Da Huo of the Xisha Marine Science Comprehensive Experimental Station, South China Sea Institute of Oceanology, Chinese Academy of Sciences for their assistance. We thank Xisha Marine Environment National Observation and Research Station. We thank BoRong Peng of National Museum of Marine Biology and Aquarium for his advice. We thank the MS center and Fan Wu of Institute of Drug Discovery Technology, Ningbo University.
Supplementary Materials
The following are available online https://www.mdpi.com/article/10.3390/md19060294/s1. Table S1 Antibacterial and antiviral activity of the isolated compounds 1–13; Figures S1–S2 Calculated and experimental ECD spectra of compounds 4 and 10; Figures S3–S81 HRESIMS, 1D and 2D NMR spectra of all new compounds 1–10; Figures S82–S83 1H-NMR for (S)-MTPA and (R)-MTPA esters of compound 5; Figures S84–S89 1D NMR spectra of known compounds 11–13.
Author Contributions
X.Y. (Xia Yan) performed isolation, structure determination of the compounds and wrote the manuscript. J.L. performed the extraction of the sample. S.H. and T.L. collected the soft coral by scuba. S.H., B.W. and X.Y. (Xiaojun Yan) revised the manuscript. H.O. performed the cytotoxic, anti-inflammatory and antimicrobial bioassays. W.W. performed the antiviral bioassays. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (2018YFC0310900), Ningbo Municipal Natural Science Foundation (2019A610201), and the Li Dak Sum Yip Yio Chin Kenneth Li Marine Biopharmaceutical Development Fund.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the Supplementary Materials file associate with this article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Haefner B. Drugs from the deep: Marine natural products as drug candidates. Drug Discov. Today. 2003;8:536–544. doi: 10.1016/S1359-6446(03)02713-2. [DOI] [PubMed] [Google Scholar]
- 2.Newman D.J., Cragg G.M. Marine natural products and related compounds in clinical and advanced preclinical trials. J. Nat. Prod. 2004;67:1216–1238. doi: 10.1021/np040031y. [DOI] [PubMed] [Google Scholar]
- 3.Gong K.-K., Tang X.-L., Zhang G., Cheng C.-L., Zhang X.-W., Li P.-L., Li G.-Q. Polyhydroxylated steroids from the South China Sea soft coral Sarcophyton sp. and their cytotoxic and antiviral activities. Mar. Drugs. 2013;11:4788–4798. doi: 10.3390/md11124788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang L.-F., Chen W.-T., Mollo E., Yao L.-G., Wang H.-Y., Xiao W., Guo Y.-W. Sarcophytrols G–L, novel minor metabolic components from South China Sea soft coral Sarcophyton trocheliophorum Marenzeller. Chem. Biodivers. 2017;14:1700079. doi: 10.1002/cbdv.201700079. [DOI] [PubMed] [Google Scholar]
- 5.Wang Q., Tang X., Liu H., Luo X., Sung P.J., Li P., Li G. Clavukoellians G–K, new nardosinane and aristolane sesquiterpenoids with angiogenesis promoting activity from the marine soft coral Lemnalia sp. Mar. Drugs. 2020;18:171. doi: 10.3390/md18030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu M.-J., Wang H., Jiang C.-S., Guo Y.-W. New cembrane-type diterpenoids from the South China Sea soft coral Sinularia crassa and their α-glucosidase inhibitory activity. Bioorganic Chem. 2020;104:104281. doi: 10.1016/j.bioorg.2020.104281. [DOI] [PubMed] [Google Scholar]
- 7.Wu Q., Sun J., Chen J., Zhang H., Guo Y.-W., Wang H. Terpenoids from marine soft coral of the genus Lemnalia: Chemistry and biological activities. Mar. Drugs. 2018;16:320. doi: 10.3390/md16090320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Q., Ye F., Li X.-L., Liang L.-F., Sun J., Sun H., Guo Y.-W., Wang H. Uncommon polyoxygenated sesquiterpenoids from South China Sea soft coral Lemnalia flava. J. Org. Chem. 2019;84:3083–3092. doi: 10.1021/acs.joc.8b02912. [DOI] [PubMed] [Google Scholar]
- 9.Izac R.R., Fenical W., Tagle B., Clardy J. Neolemnane and eremophilane sesquiterpenoids from the pacific soft coral lemnalia africana. Tetrahedron. 1981;37:2569–2573. doi: 10.1016/S0040-4020(01)98958-6. [DOI] [Google Scholar]
- 10.Yang F., Li S.-W., Zhang J., Liang L.-F., Lu Y.-H., Guo Y.-W. Uncommon nornardosinane, seconeolemnane and related sesquiterpenoids from Xisha soft coral Litophyton nigrum. Bioorganic Chem. 2020;96:103636. doi: 10.1016/j.bioorg.2020.103636. [DOI] [PubMed] [Google Scholar]
- 11.El-Gamal A.A.H., Chiu E.P., Li C.-H., Cheng S.-Y., Dai C.-F., Duh C.-Y. Sesquiterpenoids and norsesquiterpenoids from the formosan soft coral Lemnalia laevis. J. Nat. Prod. 2005;68:1749–1753. doi: 10.1021/np050326r. [DOI] [PubMed] [Google Scholar]
- 12.Zhang M., Long K., Wu H., Ma K. A novel diterpene glycoside from the soft coral of Lemnalia bournei. J. Nat. Prod. 1994;57:155–160. doi: 10.1021/np50103a024. [DOI] [Google Scholar]
- 13.Cao F., Shao C.-L., Liu Y.-F., Zhu H.-J., Wang C.-Y. Cytotoxic serrulatane-type diterpenoids from the gorgonian Euplexaura sp. and their absolute configurations by vibrational circular dichroism. Sci. Rep. 2017;7:12548. doi: 10.1038/s41598-017-12841-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anjaneyulu A.S.R., Krishnamurthy M.V.R., Rao G.V. Rare aromadendrane diterpenoids from a new soft coral species of Sinularia genus of the Indian Ocean. Tetrahedron. 1997;53:9301–9312. doi: 10.1016/S0040-4020(97)00584-X. [DOI] [Google Scholar]
- 15.Wu Z., Xie Z., Wu M., Li X., Li W., Ding W., She Z., Li C. New antimicrobial cyclopentenones from Nigrospora sphaerica ZMT05, a fungus derived from Oxya chinensis Thunber. J. Agric. Food Chem. 2018;66:5368–5372. doi: 10.1021/acs.jafc.8b01376. [DOI] [PubMed] [Google Scholar]
- 16.Lodewyk M.W., Siebert M.R., Tantillo D.J. Computational prediction of 1H and 13C chemical shifts: A useful tool for natural product, mechanistic, and synthetic organic chemistry. Chem. Rev. 2012;112:1839–1862. doi: 10.1021/cr200106v. [DOI] [PubMed] [Google Scholar]
- 17.Li G., Li H., Zhang Q., Yang M., Gu Y.-C., Liang L.-F., Tang W., Guo Y.-W. Rare cembranoids from Chinese soft coral Sarcophyton ehrenbergi: Structural and stereochemical studies. J. Org. Chem. 2019;84:5091–5098. doi: 10.1021/acs.joc.9b00030. [DOI] [PubMed] [Google Scholar]
- 18.Appendino G., Gibbons S., Giana A., Pagani A., Grassi G., Stavri M., Smith E., Rahman M.M. Antibacterial cannabinoids from Cannabis sativa: A structure-activity study. J. Nat. Prod. 2008;71:1427–1430. doi: 10.1021/np8002673. [DOI] [PubMed] [Google Scholar]
- 19.Hoffmann A., Richter M., von Grafenstein S., Walther E., Xu Z., Schumann L., Grienke U., Mair C.E., Kramer C., Rollinger J.M., et al. Discovery and characterization of diazenylaryl sulfonic acids as inhibitors of viral and bacterial neuraminidases. Front. Microbiol. 2017;8:205. doi: 10.3389/fmicb.2017.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in the Supplementary Materials file associate with this article.