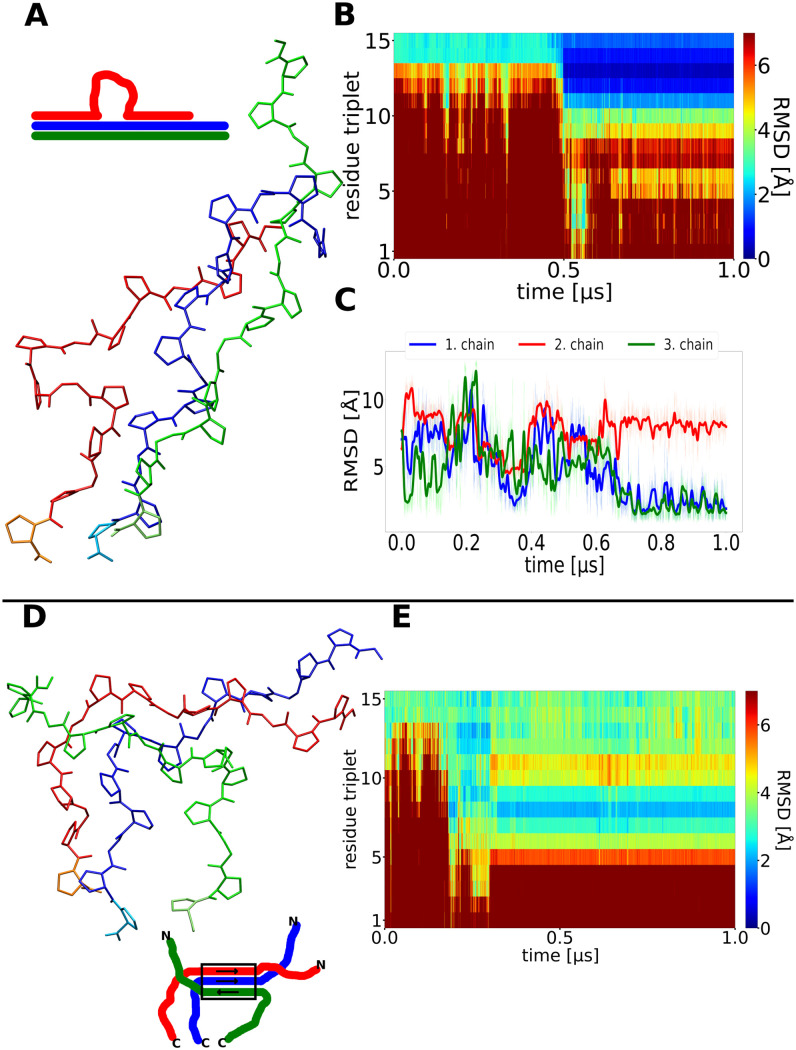
Fig 6. Misfolding of collagen triple helices during simulations.
(A) Snapshot of a finally sampled conformation with one chain (red) forming a loop but near-native conformation of the neighboring segments observed in a simulation that did not reach the native fold. (B) Time evolution of the RMSD of triplets along the simulation, after 0.5μs a correct folding of a first segment up to the loop is observed (blue regime in the plot). (C) RMSD with respect to the native folded triple helix of each chain vs. simulation time. (D) Snapshot at the final stage of a simulation with very weak restraints to stabilize the initial folding nucleus at the C-terminus of the triple helix. The green chain has detached from the other chains at the C-terminus and binds to the other two chains in a non-native arrangement. (E) The non-native arrangement remains stable for the rest of the simulation (after ~0.4 μs).