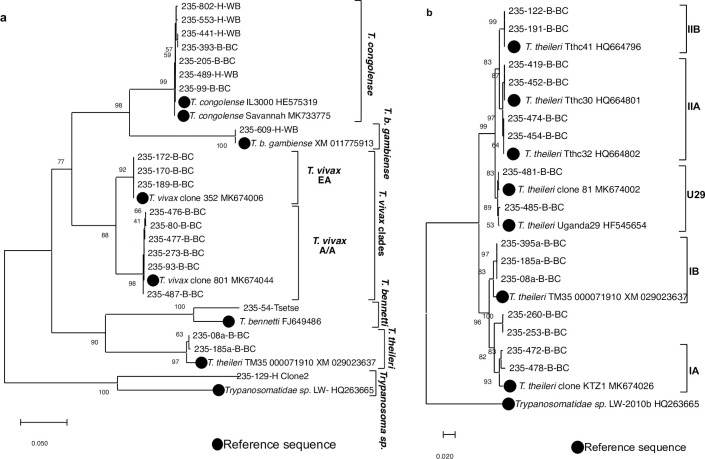
Fig 6. Neighbour-Joining trees based on alignments of gGAPDH sequences from trypanosome species detected in human, cattle, and tsetse in Southern Chad.
They were calculated using complete gap deletion and tested with 700 bootstrap replications using MEGA X software (Kumar et al., 2018). a- gGAPDH nucleotide sequences of 21 representatives of different trypanosome species and 8 reference sequences retrieved from GenBank were aligned. b- gGAPDH nucleotide sequences of 15 representatives of T. theileri clades detected only in cattle samples and 8 reference sequences belonging to IA, IB, IIA, IIB and U29 T. theileri clades retrieved from GenBank were aligned. Evolutionary analyses involved 613 bp (a) and 563 bp (b) stretches. Abbreviations: EA, East Africa; A/A: Africa/America.