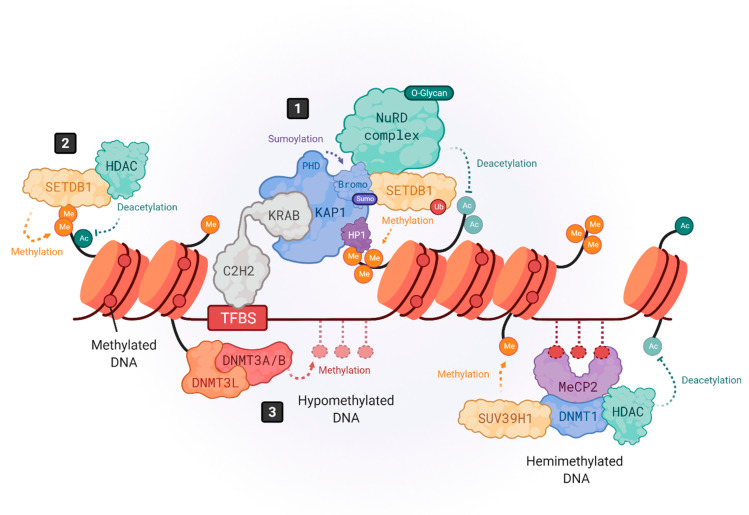
Figure 1.
Epigenetic silencing mechanisms of HERVs. The expression of HERVs is mainly controlled by DNA methylation and histone tail modifications. Histone modifications are driven by KRAB-containing zinc finger proteins (KRAB-ZFPs), which bind to transcription factor binding site (TFBS) within HERV elements, recruiting the corepressor KRAB-associated protein 1 (KAP1), also known as TRIM28 (1) [52,54]. KAP1, in turn, serves as a scaffold molecule for the assembly of heterochromatin modifiers like the histone methyltransferase SETDB1 (also known as ESET) [57], the heterochromatin protein 1 (HP1) [58] or the NuRD histone deacetylase complex [69]. Moreover, SETDB1 can also bind in a KAP1-independent way to H3K9me/K14ac marks to drive histone methylation (2) [64]. DNA methylation is driven by DNA methyltransferases (DNMTs). DNMT3L binds to histone 3 tails and recruits DNMT3A and DNMT3B to establish de novo methylation in hypomethylated DNA (3), whereas hemimethylated DNA is bound by methyl-binding proteins like MeCP2, which recruits repressor complexes such as DNMT1, histone deacetylases (HDACs) and methyltransferases like SUV39H1 to stabilize the chromatin structure repressing the gene expression (4) [47,72,73,74]. Created with Biorender.com (accessed on 21 May 2021).