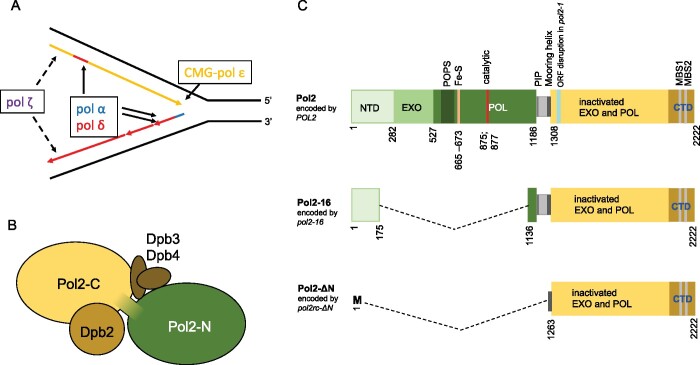
Figure 1.
DNA polymerase ε in replication in eukaryotes. (A) Current view on roles of B-family pols in replication (Guilliam and Yeeles 2020b). CMG complex travels along leading strand and unwinds DNA. Pol ε, a part of CMG, synthesizes most of the leading DNA strand. Primase/pol α and pol δ synthesize lagging strand and almost 20% of the leading strand. Pol ζ assists replication when it is impeded by DNA damage, unusual DNA structures, or impairment of the regular replication machinery (Northam et al. 2006, 2010; Garbacz et al. 2015). (B) Subunit structure of yeast pol ε. The catalytic subunit consists of two halves, active N-terminal pol (green) and inactive C-terminal pol (yellow). The second subunit, Dpb2, is indispensable. Two small subunits, Dpb3 and Dpb4, have a role in the organization of the holoenzyme (rigidity of the complex of two domains of Pol2) (Yuan et al. 2020) but are dispensable although their absence causes genome instability (Aksenova et al. 2010). (C) Top: primary structure of Pol2 protein with the location of critical elements. NTD (N-terminal domain) of unknown function, exonuclease domain, and pol domain of the active N-terminal half are shown in shades of green. Several regions critical for the function are marked. Dark green: POPS (POl2 family-specific catalytic core Peripheral Subdomain) is necessary for proper replication timing and fork progression (Meng et al. 2020), yellow bar: Fe-S cluster is necessary for pol activity (Ter Beek et al. 2019) as well as catalytic carboxylates (red) (Dua et al. 1999). The linker between two halves (gray) has a PCNA binding motif (PIP, thin black line) required for efficient stimulation of Pol ε by PCNA (Devbhandari and Remus 2020) and residues 1270-1308 that form mooring helix for binding Dpb3 and Dpb4 (Yuan et al. 2020). The yellow rectangle is the inactive C-terminal pol with metal-binding sites in the CTD. Teal bar is the site of integration of the URA3 gene in a pol2-1 allele that is viable but confers slow growth phenotype (Morrison et al. 1990). Middle: Pol2-16 protein encoded by allele pol2-16 (Kesti et al. 1999) lacks elements necessary for exonuclease and polymerase activity but possess part of the NTDand elements necessary for interaction with Dpb2, Dpb3, and Dpb4 subunits and PCNA (Ohya et al. 2002). The other name of this allele is pol2-M (Dua et al. 1999). Bottom: Pol2-ΔN encoded by pol2rc-ΔN lacks the NTD and PIP (PCNA-interaction protein box) motif but retains amino acids that can form mooring helix and C-terminal part and thus possesses the ability to interact with Dpb3 and Dpb4 subunits (Devbhandari and Remus 2020), Dpb2 and CMG (Zhou et al. 2017). The allele is also called pol2-Δ1262 or pol2-Δcat (Yeeles et al. 2017; Devbhandari and Remus 2020).