Abstract
DNA polymerases catalyze DNA synthesis through a stepwise kinetic mechanism that begins with binding to DNA, followed by selection, binding, and incorporation of a nucleotide into an elongating primer. It is hypothesized that subtle active site adjustments in a polymerase to align reactive moieties limit the rate of correct nucleotide incorporation. DNA damage can impede this process for many DNA polymerases, causing replication fork stalling, genetic mutations, and potentially cell death. However, specialized Y-family DNA polymerases are structurally evolved to efficiently bypass DNA damage in vivo, albeit at the expense of replication fidelity. Dpo4, a model Y-family polymerase from Sulfolobus solfataricus, has been well-studied kinetically, structurally, and computationally, which yielded a mechanistic understanding of how the Y- family DNA polymerases achieve their unique catalytic properties. We previously employed a real-time Förster resonance energy transfer (FRET) technique to characterize the global conformational motions of Dpo4 during DNA binding as well as nucleotide binding and incorporation by monitoring changes in distance between sites on the polymerase and DNA, and even between domains of Dpo4. Here, we extend the utility of our FRET methodology to observe conformational transitions within individual domains of Dpo4 during DNA binding and nucleotide incorporation. The results of this novel, intradomain FRET approach unify findings from many studies to fully clarify the complex DNA binding mechanism of Dpo4. Furthermore, intradomain motions in the Finger domain during nucleotide binding and incorporation, for the first time, report on the rate-limiting step of a single-nucleotide addition catalyzed by Dpo4.
Graphical Abstract
Since the initial identification of Y-family DNA polymerases,1 extensive effort has been spent to understand their precise role in DNA replication. In contrast to highfidelity, processive polymerases from A-, B-, and C-families, members of the Y-family exhibit low-fidelity, distributive DNA synthesis on undamaged DNA.2 Though these are undesirable traits for a replicative DNA polymerase, they uniquely qualify the Y-family polymerases for the critical cellular responsibilities of immunoglobulin generation, somatic hypermutation, and translesion DNA synthesis (TLS).3 Perhaps the most striking role of the Y-family polymerases involves their abilities to bypass DNA damage during TLS, which would otherwise stall DNA synthesis by high-fidelity replicative DNA polymerases and lead to cell death. Much of what is currently known concerning the mechanism of DNA synthesis on undamaged and damaged DNA by the Y-family DNA polymerases comes from structural4–6 and kinetic2,7–10 investigations of Dpo4, the model and lone Y-family DNA polymerase from Sulfolobus solfataricus.
Akin to other Y-family members, Dpo4 contains a structurally conserved polymerase core consisting of Finger, Palm, and Thumb domains arranged in a characteristic right-hand architecture, as well as a Little Finger (LF) domain that is joined to the polymerase core through a highly flexible peptide linker (Figure 1).4 Interestingly, there is some evidence to suggest that the LF domain and linker mediate the association of Dpo4 with DNA11 and that the LF domain determines the lesion bypass properties of Y-family polymerases.12 In contrast to many other polymerases, structural comparison of the binary (Dpo4•DNA) and ternary (Dpo4•DNA•dNTP) complexes of the enzyme reveals that Dpo4 does not exhibit the open to closed conformational transition of the Finger domain upon nucleotide binding thought to function as a fidelity checkpoint but rather exhibits a largely preformed active site lacking any major conformational adjustments.6 Nevertheless, ensemble13–17 and single-molecule18,19 techniques have revealed Dpo4 to be dynamic, exhibiting rigid body domain movements as well as more local conformational motions throughout DNA association, nucleotide binding, and nucleotide incorporation steps of catalysis, and these motions were found to be altered by the presence of a lesion in the DNA template.14,19
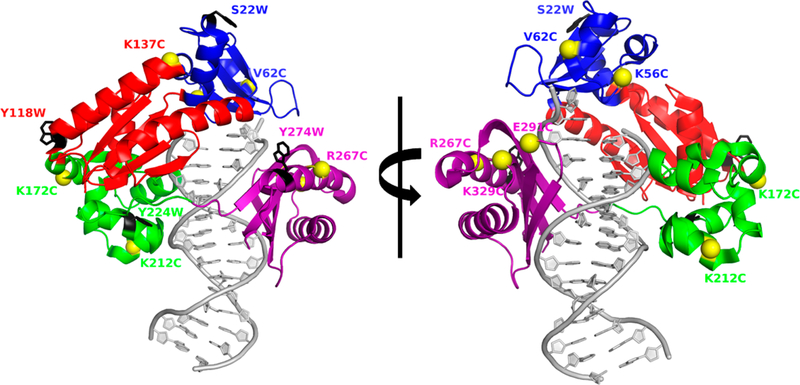
Figure 1.
Locations of intradomain donor and acceptor FRET pairs mapped onto the X-ray crystal structure of DNA-bound Dpo4. The Finger, Palm, Thumb, and LF domains are colored blue, red, green, and purple, respectively. Each domain contains one, unique Trp donor residue (black stick models) and one or more sites for CPM acceptor attachment (yellow spheres). The DNA substrate is colored gray. For the sake of clarity, the right structure is rotated 180° relative to the left structure.
Pre-steady-state kinetic characterization of the minimal kinetic mechanism of nucleotide insertion by Dpo4 suggests that a conformational change, likely consisting of subtle active site rearrangements to properly align divalent metal ions and carboxylate moieties as well as reactive groups of the primer and incoming nucleotide, is rate-limiting, rather than phosphodiester bond formation.7 Unfortunately, there is no explicit evidence of this conformational transition, leading to significant controversy.20 For A-, B-, and C-family and some X-family polymerases, it was thought that the closure of the Finger domain during nucleotide binding may signify the slowest step during single-nucleotide incorporation,21,22 but structural studies indicate that Dpo4 lacks such a change. Our previous, stopped-flow Förster resonance energy transfer (FRET) study 13 sought to capture the rate-limiting conformational change for Dpo4 but instead revealed global dynamic conformational events at several other stages during nucleotide binding and incorporation.
Here, we extended our stopped-flow FRET methodology to observe conformational motions that may exist within individual domains of Dpo4 during DNA and nucleotide binding and catalysis. By generating intradomain FRET constructs consisting of donor and acceptor fluorophores within each domain of Dpo4, we were able to gain significant insight into the DNA binding mechanism of Dpo4 and observe dynamic motions within each domain during nucleotide binding and insertion. The existence of slow conformational motions observed for the Finger domain was independent of phosphodiester bond formation, and the motions may be reporting on the elusive rate-limiting step for single-nucleotide incorporation.
MATERIALS AND METHODS
Preparation of Protein and DNA
FRET constructs were generated as previously described.15 Briefly, donor Trp residues were individually introduced into each domain of Dpo4. For each Trp-containing mutant, a corresponding Cys mutation was generated within the same domain for subsequent labeling with the acceptor fluorophore, 7-(diethylamino)-3-(4′-maleimidyl-(phenyl)-4-methylcoumarin (CPM) (ThermoFisher Scientific). Notably, Dpo4 contains no native Trp residues, and the lone native Cys residue was mutated to Ser. All mutations were confirmed by DNA sequencing and are shown in Figure 1. Expression, purification, and fluorophore labeling were completed as previously described.15 Unreacted CPM dye was removed by two consecutive spins through Micro P-6 BioSpin columns (Bio-Rad). The protein concentration was determined using the Bradford assay (Bio-Rad) with wild-type assessed by measuring the concentration of CPM by UV/vis spectrophotometry using an extinction coefficient of 44800 M−1 cm−1 at 430 nm. For all CPM-labeled mutants, the ratio of dye concentration to protein concentration revealed a labeling efficiency of ≥90%.
DNA oligonucleotides were purchased from Integrated DNA Technologies (IDT). A 21-mer primer (5′-CGAGCCGTCGCATCCTACCGC-3′) with either a normal, 3′-deoxy-terminated end or a 3′-dideoxy-terminated end was annealed to a 30-mer template (5′-GATGCTGCAGCGGTAGGATGCGACGGCTCG-3′) to generate the DNAOH or DNAH substrate, respectively. All synthetic DNA oligonucleotides were purified by denaturing polyacrylamide gel electrophoresis (PAGE) (17% polyacrylamide, 8 M urea), and the concentrations were determined by the UV absorbance at 260 nm. Alexa488-labeled DNAOH was generated as described previously13,14 through Alexa Fluor 488-NHS ester (Invitrogen) labeling of a 5-C6-amino-2′-deoxythymidine modification (T) on the ninth base from the primer 3′ terminus.
Buffers
Unless otherwise specified, all reactions were performed in buffer R, containing 50 mM HEPES (pH 7.5 at 20 °C), 6 mM MgCl2, 50 mM NaCl, 0.1 mM EDTA, 1 mM DTT, and 10% glycerol.
Steady-State Fluorescence Spectroscopy Assays
All steady-state fluorescence spectra were recorded on a Fluoromax-4 fluorometer (Jobin Yvon Horiba) at 20 °C. The slit widths for the excitation and emission monochromators were each set to 5 nm. Fluorescence emission spectra of each CPM-labeled Dpo4 mutant (200 nM) in the absence or presence of either DNA (DNAOH, 300 nM) or DNA (300 nM) and the correct nucleotide (dTTP, 1 mM) were recorded upon Trp donor excitation at 290 nm. Upon addition of DNA and dTTP, a 5 min incubation period was observed to allow for reaction equilibration. All reported spectra were corrected for dilution and for the intrinsic fluorescence of buffer components as well as of DNAOH and dTTP, when applicable.
Notably, while the distance between donor and acceptor probes can be calculated from the FRET efficiency, this requires consideration of many complex variables.23,24 Therefore, to simplify our data analysis, we have calculated (ratio) by dividing the acceptor fluorescence due to energy transfer from the excited donor fluorophore by the acceptor fluorescence due to direct excitation. Importantly, changes in (ratio)A observed with a change in experimental conditions are indicative of conformational change as (ratio)A is directly proportional to FRET efficiency.
Stopped-Flow FRET Assays
Stopped-flow fluorescence spectroscopy experiments were completed at 20 °C using an Applied Photophysics SX20 instrument with a dead time of ∼1 ms and monochromator slit widths set to 5 nm. Importantly, a temperature of 20 °C was maintained for all stopped-flow mixing experiments to be consistent with our previous studies and to permit observation of rapid fluorescent phases that may become too fast to be detected at elevated temperatures.13–15 During excitation at 290 nm, reaction components (all reported component concentrations represent final concentrations after mixing) were rapidly mixed within the 20 μL optical cell, and subsequent Trp or CPM fluorescence was recorded over time using a 305 or 420 nm cutoff filter (Applied Photophysics Ltd.), respectively. The resulting fluorescence traces were fit by nonlinear regression using Pro-Data Viewer software (Applied Photophysics Ltd.) to either single-exponential (eq 1), double-exponential (eq 2), or triple-exponential (eq 3) equations (shown below) depending on the number of observed FRET phases.
| (1) |
| (2) |
| (3) |
where F(t) signifies the change in fluorescence over time (t); A, A1, A2, and A3 are the phase amplitudes; and kobs, k1, k2, and k3 are the corresponding phase rates. All reported rates and errors represent the average rates and corresponding standard deviations among multiple, replicate stopped-flow mixing experiments.
Stopped-Flow Anisotropy Assays
Following the manufacturer’s protocol, stopped-flow anisotropy measurements were performed using the Applied Photophysics SX20 instrument at 20 °C outfitted with a T-format fluorescence polarization accessory (Applied Photophysics Ltd.) and monochromator slit widths set to 10 nm. For experiments monitoring the binding of wt Dpo4 (500 nM) to Alexa488-labeled DNAOH (500 nM), polarized excitation occurred at 499 nm, and 530 nm cutoff filters were placed in front of each detector. Kinetic parameters from anisotropy measurements were obtained by fitting the data to a single-exponential equation (eq 1).
Dpo4 Mutant Activity Assays
Enzymatic activities of Dpo4 mutants following CPM labeling were assessed at 20 °C using a burst kinetic assay, as described previously.19,25 For each CPM-labeled mutant and wt Dpo4, 60 nM 5′−32P-labeled DNAOH was preincubated in buffer R with 10 nM enzyme before being rapidly mixed with 100 μM dTTP using a rapid chemical-quench-flow apparatus (Kintek). Reactions were then quenched at increasing time intervals with the addition of 0.37 M EDTA. Products were then separated by denaturing PAGE (17% polyacrylamide, 8 M urea, 1× TBE running buffer) and quantitated by phosphorimaging (Typhoon Trio, GE Healthcare). Product formation was plotted against time and fit by nonlinear regression using KaleidaGraph (Synergy Software) to the burst equation:
| (4) |
where [P](t) signifies the change in product concentration over time (t), A is the burst phase amplitude, kburst is the single-turnover nucleotide incorporation rate constant, and kss is the steady-state rate constant.
RESULTS
Design of an Intradomain FRET System
To detect movements within the individual domains of Dpo4, we modified our previously described FRET system 13,15 in which a single, site-specifically engineered Trp residue serves as the donor fluorophore, and CPM, conjugated to a single, site-specifically engineered Cys residue, serves as the acceptor fluorophore (Figure 1 and Table 1). When selecting sites for acceptor labeling, we strove (1) to mutate only surface-exposed amino acid residues and (2) to optimize the distance to the Trp donor, based on the Trp–CPM Förster radius (R0) of 30 Å.26,27 We sought to keep the distance between donor and acceptor sites within the range of R0 ± 0.5R0 but had to compromise in several instances because of distance constraints imposed by the individual domains (Table 1). In this study, we will refer to unlabeled Dpo4 constructs by their respective Trp and Cys mutations (e.g., Y274W-E291C) and to CPM-labeled Dpo4 constructs in a similar manner but with a superscripted CPM (e.g., Y274W-E291CCPM). 32P-based pre-steady-state kinetic burst assays of wt Dpo4 and each CPM-labeled construct were performed to verify enzymatic activity (Materials and Methods). We found that neither the single-turnover nucleotide incorporation rate constant (kburst, eq 4) nor the steady-state multiple-turnover rate constant (kss, eq 4) was appreciably affected by the mutations or labeling when compared to that of wt Dpo4 (Table 2). These results suggest that our labeling procedure did not impact the polymerase activity of Dpo4.
Table 1.
Distances between Trp Donor Residues and Cys-Conjugated CPM Acceptor Fluorophores Projected from X-ray Crystal Structures
| distancea (Å) |
||||
|---|---|---|---|---|
| domain | mutant | apob | binaryc | ternaryd |
| Little Finger | Y274W-R267CCPM | 10.8 | 10.7 | 10.8 |
| Y274W-E291CCPM | 28.3 | 28.3 | 28.4 | |
| Y274W-K329CCPM | 28.6 | 28.4 | 28.4 | |
| Thumb | Y224W-K172CCPM | 16.3 | 16.2 | 16.4 |
| Y224W-K212CCPM | 14.5 | 14.8 | 14.8 | |
| Palm | Y118W-K137CCPM | 26.6 | 26.1 | 25.8 |
| Finger | S22W-K56CCPM | 22.0 | 20.9 | 21.1 |
| S22W-V62CCPM | 24.8 | 24.5 | 24.3 | |
Estimated distances are calculated from crystal structures and do not account for the flexible linker utilized to attach the CPM fluorophore to the introduced Cys residues.
Distances calculated between the Cα atoms of residues in the apo structure of Dpo4 (PDB entry 2RDI).
Distances calculated between the Cα atoms of residues in the Dpo4·DNA binary complex structure (PDB entry 2RDJ).
Distances calculated between the Cα atoms of residues in the Dpo4·dideoxy-DNA·dATP ternary complex structure (PDB entry 2AGQ).
Table 2.
Kinetic Parameters of wt Dpo4 and Each CPM-Labeled Dpo4 Mutant Measured Using the Pre-Steady-State Burst Assay at 20 °C
| enzyme | kbursta (s−1) | kssb (s−1) |
|---|---|---|
| wild-type Dpo4 | 0.5 ± 0.1 | 0.007 ± 0.001 |
| Y274W-R267CCPM | 0.6 ± 0.1 | 0.004 ± 0.001 |
| Y274W-E291CCPM | 0.6 ± 0.1 | 0.006 ± 0.001 |
| Y274W-K329CCPM | 0.4 ± 0.2 | 0.014 ± 0.003 |
| Y224W-K172CCPM | 0.30 ± 0.05 | 0.012 ± 0.001 |
| Y224W-K212CCPM | 0.3 ± 0.2 | 0.013 ± 0.003 |
| Y118W-K137CCPM | 0.5 ± 0.1 | 0.007 ± 0.002 |
| S22W-K56CCPM | 0.3 ± 0.1 | 0.007 ± 0.008 |
| S22W-V62CCPM | 0.3 ± 0.2 | 0.011 ± 0.002 |
Obtained from burst kinetic assays in which a preincubated solution of a Dpo4 mutant (10 nM) and 5′-32P-labeled DNAOH (60 nM) was rapidly mixed with dTTP (100 μM) and the reaction subsequently quenched at various time points with EDTA (0.37 M). The plot of product concentration vs time was fit to the burst equation [product] = A[1 – exp(-kburstt) + ksst], where A is the burst phase amplitude, kburst is the observed burst rate, and kss is the observed steady-state rate.
Percent of the active enzyme normalized to wild-type Dpo4 as calculated from the burst phase amplitude.
Dpo4 Intradomain Motion Observed by Steady-State Fluorescence
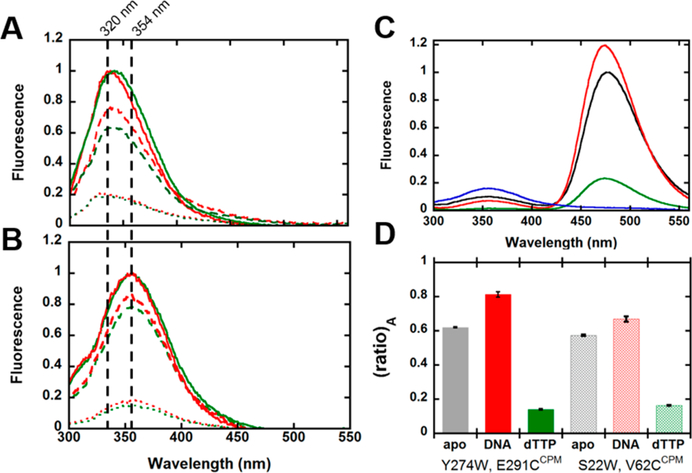
The Trp fluorescence of each Dpo4 construct prior to CPM labeling was investigated by steady-state i fluorescence (Figure 2). Interestingly, we observed that Trp s residues introduced into the Palm (Y118W) and Thumb (Y224W) domains displayed maximal fluorescence emission at 320 nm (Figure 2A) while Trp residues introduced into the; Finger (S22W) and LF (Y274W) domains displayed a red-shifted maximal fluorescence emission at 354 nm, suggesting a higher degree of solvent exposure (Figure 2B).6,24,28 These results qualitatively agree with our expectations based on their locations on Dpo4 in the DNA-bound state presented in Figure 1, compared to the apo state.6 The addition of 300 nM DNAOH (Materials and Methods) resulted in a 16–35% decrease in fluorescence for all Trp mutants studied. The subsequent addition of 1 mM dTTP resulted in an additional 34–49% decrease in the intensity of the fluorescence signal. Unfortunately, given that both DNA and nucleotide absorb at 290 nm, significant excitation light is likely being lost to inner filtering, thereby contributing to the decreases in the intensity of the fluorescent signal. However, we would anticipate that if the decreasing signals were due solely to inner filtering then a uniform reduction in the intensity of the signal would be observed for all mutants. Instead, we see signal attenuation to varying degrees among the tested Trp mutants, implying protein conformational dynamics upon the addition of DNA and nucleotide.
Figure 2.
Steady-state fluorescence of Trp-containing Dpo4 mutants and select CPM-labeled intradomain FRET constructs. Emission spectra for Dpo4 mutants (200 nM) were recorded at 20 °C with an excitation wavelength of 290 nm. (A) Emission spectra of mutants Y118W-K137C (green) and Y224W-K172C (red) in the apo state (solid lines) and after sequential additions of 300 nM DNAOH (dashed lines) and 1 mM dTTP (dotted lines). (B) Emission spectra of mutants Y274W-E291C (green) and S22W-V62C (red) in the apo state (solid lines) and after sequential additions of differences in the maximal emission wavelengths among the various Trp mutants. (C) Emission spectra for CPM-labeled mutant Y274W-E291CCPM in the apo state (black), and after sequential additions of 300 nM DNAOH (red) and 1 mM dTTP (green). The emission spectrum of Y274W-E291C before CPM labeling is colored blue. (D) Differences in (ratio)A for select CPM-labeled Dpo4 mutants in the apo state and following sequential additions of 300 nM DNAOH and 1 mM dTTP. Error bars represent the standard deviation calculated from three separate experiments.
The emission of Dpo4 mutant Y274W-E291CCPM was recorded upon excitation at 290 nm and revealed a significant reduction in Trp emission (in Figure 2C, compare the blue trace with the black trace) with a concomitant increase in fluorescence at 472 nm, the emission maximum of CPM (Figure 2C, black trace). We calculated (ratio)A for select Dpo4 mutants (Figure 2D), Y274W-E291CCPM (solid bars) and S22W-V62CCPM (hashed bars), from the collected emission spectra of the apo state and following sequential additions of 300 nM DNAOH and 1 mM dTTP. As true FRET efficiency values depend on multiple complex parameters,23,24 we instead used (ratio)A, calculated as the acceptor fluorescence due to energy transfer divided by the acceptor fluorescence due to direct excitation, as a convenient indicator of the change in the distance between the donor and acceptor probes under varying experimental conditions (Materials and Methods). For Y274W-E291CCPM and S22W-V62CCPM, the addition of DNAOH resulted in 32 and 17% increases in (ratio)A, respectively, indicating that the FRET probes moved closer together for binary complex formation (Dpo4•DNAOH). In contrast, the subsequent addition of 1 mM dTTP resulted in a significant decrease in (ratio)A for Y274W-E291CCPM (82%) and S22W-V62CCPM (76%). As dTTP strongly absorbs at 290 nm, the dramatic reductions in (ratio)A, as a result of concurrent decreases in Trp and CPM fluorescence intensities, for both Dpo4 mutants likely stem from the inner filtering caused by the high concentration of dTTP (1 mM) needed to saturate nucleotide binding, rather than from nucleotide-dependent conformational dynamics. Despite the significant inner filtering, the difference in signal attenuation upon addition of dTTP may still indicate changes in distance between the donor and acceptor probes as inner filtering should attenuate the total fluorescence to the same degree for each mutant. Steady-state fluorescence spectra for the other Dpo4 mutants were also recorded and produced results (data not shown) consistent with the expected distance changes determined from the crystal structures (Table 1). These experiments were repeated with the remaining CPM-labeled Dpo4 mutants listed in Table 1, and the anticorrelated signal changes were observed for each to clearly report on the efficient energy transfer occurring from the Trp donor to the CPM acceptor (data not shown). Taken together, these results suggest that conformational dynamics within individual polymerase domains are taking place during substrate binding and catalysis.
Conformational Dynamics of Intradomain Little Finger Mutants during DNA Binding
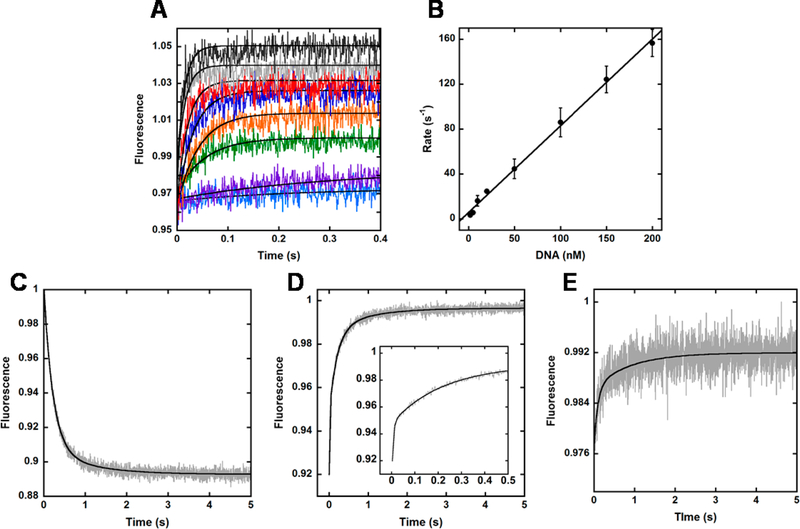
To further study the DNA binding process of Dpo4, we monitored fluorescence changes upon rapidly mixing each of the intradomain FRET constructs with DNAOH in a stopped-flow apparatus at 20 °C. Briefly, 100 nM CPM-labeled Dpo4 mutant was rapidly mixed with 100 nM DNAOH in the stopped-flow apparatus, and the fluorescence emission of CPM upon excitation at 290 nm was monitored using a 420 nm cutoff filter. For several of the Dpo4 mutants, including Y118W-K137CCPM, Y224W-K172CCPM, and Y224W-K212CCPM, time-dependent decreases in fluorescence with low signal-to-noise ratios were observed upon mixing with DNAOH (Figure S1). This result suggests that the conformational movements upon DNA binding in the Palm and Thumb domains may not be sufficiently large in magnitude to cause a significant fluorescence signal change. However, experiments repeated with LF domain mutants Y274W-K329CCPM, Y274W-E291CCPM, and Y274W-R267CCPM, as well as Finger mutants S22W-K56CCPM and S22W-V62CCPM, demonstrated time-dependent increases in CPM fluorescence with high signal-to-noise ratios upon mixing with DNAOH (Figure S2). To gain insight into the conformational changes observed for the LF domain mutants, we monitored the fluorescence response of Y274W-K329CCPM upon rapid mixing with varying concentrations of DNAOH (2–200 nM) (Figure 3). To ensure an adequate signal change while maintaining pseudo-first-order reaction conditions of excess polymerase, 100 nM Y274W-K329CCPM was used for lower concentrations of DNAOH (2–20 nM) and 1 μM Y274W-K329CCPM was used for higher concentrations of DNAOH (50–200 nM). Interestingly, we found that the observed rates of fluorescence change increased linearly with increasing concentrations of DNAOH (Figure 3A). By plotting the observed pseudo-first-order rate constants versus their corresponding DNA concentrations, we were able to extract the second-order, bimolecular association rate constant (kon) of 7.8 × 108 M−1 s−1, which suggests that initial binary complex formation is nearly diffusion-limited (Figure 3B).29 We performed similar, stopped-flow DNA titration experiments with LF domain mutant Y274W-E291CCPM and obtained comparable results (Figure S3A).
Figure 3.
Conformational changes in the LF domain upon DNA binding. (A) Dpo4 LF domain mutant Y274W-K329CCPM (100 or 1000 nM) was rapidly mixed with varying concentrations of DNAOH (from 2 to 200 nM, from light blue to black traces, respectively), and the CPM fluorescence was monitored upon excitation at 290 nm. The black lines depict single-exponential fits to the data. (B) The observed rate constants (kobs) extracted from panel A were plotted against the corresponding DNA concentrations. Errors bars represent the standard deviations calculated from multiple replicate experiments. The slope of the best-fit line to the data signifies the bimolecular association rate constant (kon) for binding of Dpo4 to DNAOH (7.8 × 109 M−1 s−1). (C) CPM fluorescence ofY274W-K329CCPM (100 nM) preincubated with DNAOH (100 nM) upon excitation at 290 nm following rapid mixing with wt Dpo4 trap (2 μM). (D) CPM fluorescence of Y274W-K329CCPM (2 μM) upon excitation at 290 nm following rapid mixing with a preincubated solution of wt Dpo4 (100 nM) and DNAOH (100 nM). The inset features the fast time points to illustrate the multiexponential behavior of the data. (E) Trp fluorescence of Y274W-K329C (100 nM) preincubated with DNAOH (100 nM) upon excitation at 290 nm following rapid mixing with wt Dpo4 trap (2 μM) The black lines depict double-exponential (C and E) or triple-exponential (D) fits to the data.
To study the reverse conformational change of the LF domain upon dissociation of the binary complex (Dpo4•DNAOH), we performed several trap assays (Figure 3C–E). A preincubated solution of Y274W-K329CCPM (100 nM) and DNAOH (100 nM) was rapidly mixed with a 20-fold excess of wt Dpo4 (2 μM), which contains no native Trp residues, and the fluorescence of CPM was monitored upon excitation at 290 nm (Figure 3C). The resulting fluorescence decrease was fit to a double-exponential equation (Materials and Methods) to yield a fast phase rate of 4.5 ± 0.2 s−1 and a slow phase rate of 1.00 ± 0.09 s−1. As an alternative approach to measuring the reverse conformational change, the CPM fluorescence upon excitation at 290 nm was monitored for a preincubated solution of wt Dpo4 (100 nM) and DNAOH (100 nM) following rapid mixing with a 20-fold excess of Y274W-K329CCPM (2 μM). The resulting fluorescence trace exhibited multiexponential kinetic behavior, displaying an initial fast phase increase with a rate of 169 ± 20 s−1, followed by a second phase with a rate of 5 ± 1s−1 and a third, slow phase with a rate of 0.8 ± 0.4 s−1. The two slower phase rates are in agreement with the previous trap assay. While it is not well resolved, we suspect that the initial fast phase represents the population of DNAOH that remained unbound by wt Dpo4 following preincubation and was rapidly bound by the fluorescent Y274W-K329CCPM trap upon mixing. Given that Dpo4 binds to DNA with an equilibrium dissociation constant (KD) of ∼10 nM,7,10,19 we would estimate that ∼70% of the DNAOH would be prebound by wt Dpo4, which would leave ∼30 nM unbound DNAOH. Consistently, the observed fast phase amplitude (42 ± 7%) suggests that ∼40 nM DNAOH remained unbound prior to mixing, and the corresponding fast rate (169 ± 20 s−1) was not accurately measured on the time scale of data acquisition, as we would expect for mixing with a high concentration of Y274W-K329CCPM (2 μM) (Figure 3B). As the association of Y274W-K329CCPM with DNAOH was limited by the dissociation of wt Dpo4, the remaining slow rates (5 ± 1 and 0.8 ± 0.4 s−1) describe the LF intradomain reverse conformational change upon binary complex dissociation and are consistent with the previous trap experiment (4.5 ± 0.2 and 1.00 ± 0.09 s−1, respectively). A trap experiment measuring the Trp emission upon excitation at 290 nm of Y274W-K329C (100 nM) preincubated with DNAOH followed by rapid mixing with a 20-fold excess of wt Dpo4 (2 μM) also showed a biphasic increase in acceptor, CPM fluorescence with rates of 9 ± 2 and 0.7 ± 0.3 s−1 (Figure 3E). This result agrees with the steady-state fluorescence data of Figure 2B, where the addition of 300 nM DNAOH to apo-Y274W-K329C resulted in a 22% decrease in donor, Trp emission, as well as with the rates for the reverse conformational change upon DNA dissociation reported above (Figure 3C,D). Notably, similar trap experiments were conducted with another LF intradomain mutant, Y274W-E291CCPM, and yielded comparable results (Figure S3B,C).
Conformational Dynamics of Intradomain Finger Mutants during DNA Binding
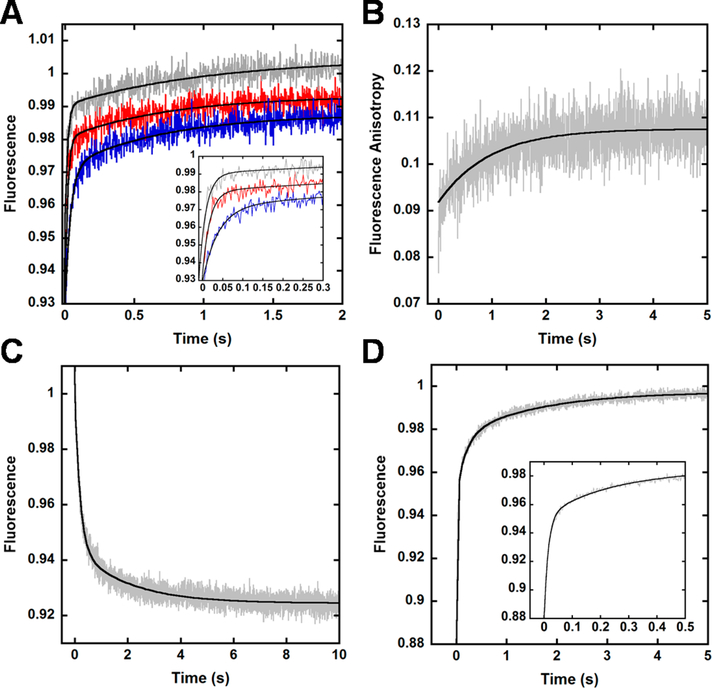
To gain insight into the conformational changes observed for the Finger domain mutants (Figure S2), we monitored the fluorescence response of S22W-V62CCPM upon rapid mixing with varying concentrations of DNAOH (20–100 nM) (Figure 4A). Interestingly, as with LF domain mutant Y274W-K329CCPM (Figure 3), we observed fluorescence changes that were dependent on the concentration of DNAOH as indicated by a comparable linear increase in rates (25 ± 3, 55 ± 11, and 90 ± 10 s−1 for 20, 50, and 100 nM, respectively) (Figure 4A and inset). However, we also detected an additional, slower fluorescence phase that was independent of DNAOH concentration with an average rate of 1.5 ± 0.3 s−1 and an amplitude of 26 ± 2% (Figure 4A). This slower phase may represent a conformational change in the Finger domain subsequent to DNA binding, or a population of Dpo4 molecules that must first undergo a change in conformation to allow for DNA binding. To distinguish between these two possibilities, we performed stopped-flow fluorescence anisotropy experiments (Materials and Methods). If a conformational change in the Finger domain were to occur subsequent to DNAOH binding, then no detectable change in fluorescence anisotropy should be observed as this likely small conformational adjustment would not be expected to appreciably perturb the tumbling of the large binary (Dpo4•DNA) complex. However, if instead a population of Dpo4 were to undergo a slow conformational adjustment before binding to DNA, we would observe a time-dependent change in anisotropy with a rate comparable to the slow phase of the previous FRET experiment (Figure 4A). Indeed, upon mixing of wt Dpo4 (500 nM) with Alexa488-labeled DNAOH (500 nM) and excitation at 499 nm, we observed a time-dependent change in anisotropy with a rate of 1.2 ± 0.2 s−1 (Figure 4B), indicating that a population of Dpo4 molecules was slowly associating with the DNA. Notably, at the reagent concentrations (500 nM Dpo4 and 500 nM DNAOH) used to achieve a sufficient anisotropy signal, the fast phase association was too rapid to be clearly detected. Control experiments in which Alexa488-labeled DNAOH was mixed with buffer showed a stable anisotropy signal over time (Figure S4A), indicating that the anisotropy changes shown in Figure 4B resulted from binding of Dpo4 to DNA. Furthermore, we attribute the lack of a readily apparent, slow fluorescence phase in the data collected for the LF domain mutants (Figure 3A and Figure S2) to poorer signal-to-noise ratios during data acquisition for those constructs. However, it is clear that at longer time scales, the DNA binding data for the LF domain constructs significantly deviate from a single-exponential fit (Figure S2A–C), thereby further substantiating the slower binding event.
Figure 4.
Conformational changes in the Finger domain upon DNA binding. (A) Dpo4 Finger mutant S22W-V62CCPM (100 nM) was rapidly mixed with varying concentrations of DNAOH (from 20 to 200 nM, from blue to gray traces, respectively), and the CPM fluorescence was monitored upon excitation at 290 nm. The black line depicts a double-exponential fit to the data, and the inset highlights the concentration dependence of the initial fast rate. (B) wt Dpo4 (500 nM) was rapidly mixed with Alexa488-labeled DNAOH (500 nM), and the change in fluorescence anisotropy was measured upon excitation at 499 nm. The black line depicts a single-exponential fit to the data. (C) CPM fluorescence of S22W-V62CCPM (100 nM) preincubated with DNAOH (100 nM) upon excitation at 290 nm following rapid mixing with a 20-fold excess of wt Dpo4 (2 μM). The black line depicts a double-exponential fit to the data. (D) CPM fluorescence of S22W-V62CCPM (2 μM) was monitored upon excitation at 290 nM following rapid mixing with a preincubated solution of wt Dpo4 (100 nM) and DNAOH (100 nM). The black line depicts a triple-exponential fit to the data, and the inset features the fast time points to illustrate the multiexponential behavior of the data.
To investigate the reverse conformational change of the Finger domain upon dissociation from DNA, we performed several trap assays as previously completed for LF domain mutant Y274W-K329CCPM. A preincubated solution of S22W-V62CCPM (100 nM) and DNAOH (100 nM) was rapidly mixed with a 20-fold excess of wt Dpo4 (2 μM), and the CPM fluorescence upon excitation at 290 nm was recorded (Figure 4C). Like Y274W-K329CCPM, S22W-V62CCPM dissociated with biphasic kinetics. The fast (5.1 ± 0.3 s−1) and slow (0.6 ± 0.1 s−1) phase rates were close to those measured for Y274W-K329CCPM (4.5 ± 0.2 and 1.00 ± 0.09 s−1, respectively). Furthermore, the results of another trap assay in which a preincubated solution of wt Dpo4 (100 nM) and DNAOH (100 nM) was rapidly mixed with S22W-V62CCPM (2, μM) and the CPM fluorescence upon excitation at 290 nm was monitored supported this result (Figure 4D). As with the LF domain mutant, the resulting fluorescence increase was best fit to a triple-exponential equation to yield an initial fast phase rate of 94 ± 8 s−1 (which is likely a result of S22W-V62CCPM rapidly associating with unbound DNAOH) and two other phase rates of 5 ± 1 and 0.5 ± 0.2 s−1, which were comparable to those acquired from Figure 4C. Moreover, the presence of a slow phase in the dissociation experiments necessitates a slow association step as observed above (Figure 4A), rather than equilibrium binding of Dpo4 (i.e., function of forward and reverse rate constants). Together with the results for the LF domain mutants, these data further illuminate the complex binding kinetics of Dpo4.
Intradomain Conformational Dynamics during Correct Nucleotide Binding and Incorporation
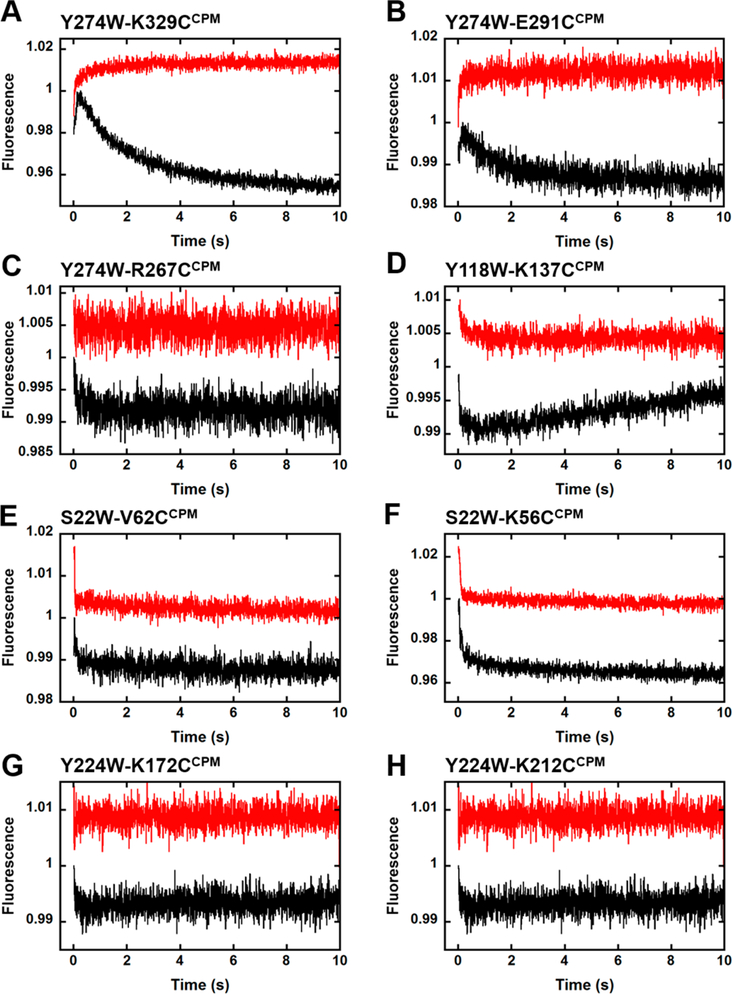
To monitor the intradomain motions of Dpo4 during correct nucleotide incorporation, CPM fluorescence was recorded upon excitation at 290 nm following rapid mixing of preincubated solutions of CPM-labeled Dpo4 mutants (200 nM) and DNAOH (300 nM) with dTTP (1 mM) (Figure 5). Notably, we performed several control experiments to verify that fluorescence changes resulted from distance changes rather than photophysical artifacts such as changes in the environment of the Trp probe or inner filtering (Figure S4). Each domain of Dpo4 undertook conformational dynamics upon mixing with nucleotide, demonstrating that in addition to altering its global domain architecture, the polymerase also employs more subtle intradomain motions to achieve catalysis. Within the LF domain, Dpo4 mutants Y274W-K329CCPM (Figure 5A) and Y274W-E291CCPM (Figure 5B) displayed biphasic fluorescence changes during nucleotide insertion consisting of a rapid [16 ± 3 and 10 ± 2 s−1, respectively (Table 3)] increase phase (P1) and a slow [0.46 ± 0.02 and 0.89 ± 0.04 s−1, respectively (Table 3)] decrease phase (P2). These data suggest that the donor and acceptor probes rapidly moved close to one another before slowly moving apart. In contrast, LF domain mutant Y274W-R267CCPM (Figure 5C) exhibited a single, rapid decrease phase (P1) [6 ± 1 s−1 (Table 3)], although with a much smaller amplitude, implying the fast separation of the FRET pair. In the Palm domain, Y118W-K137CCPM (Figure 5D) revealed a fast initial decrease phase (P1) subsequent to a slow increase phase (P2) upon nucleotide addition. Interestingly, Finger domain mutants S22W-V62CCPM (Figure 5E) and S22W-K56CCPM (Figure 5F) demonstrated domain motions similar to those of the Palm domain during nucleotide incorporation and displayed P1 and P2 rates comparable to those of other intradomain mutants (Table 3). However, the motion of P2, as observed through the Finger domain, was in the same direction as that of P1. Thus, while the majority of P2 phases occur in the opposite direction of their corresponding P1 phase, indicating a domain relaxation after catalysis, the intradomain motions of the Finger domain suggest additional gripping motions following P1. With the Thumb domain mutants Y224W-K172CCPM (Figure 5G) and Y224W-K212CCPM (Figure 5H), each displayed only a single, fast decrease in fluorescence (P1). Because of the tight distance constraints imposed by our intradomain analysis, Y274W-R267CCPM and both Thumb domain mutants were at the practical limit of our Trp-CPM FRET system with interprobe distances near or less than 0.5R0 (Table 1). Consequently, the signal-to-noise ratios for data collected with these mutants were less than ideal and could explain atypical results, including the loss of the P2 fluorescence phase, observed during nucleotide insertion within their respective domains.
Figure 5.
Intradomain conformational dynamics during nucleotide binding and incorporation. (A–H) Changes in CPM fluorescence upon excitation at 290 nm following rapid mixing of a preincubated solution of an intradomain Dpo4 mutant (200 nM) and either DNAOH (300 nM,black traces) or DNAH (300 nM, red traces) with dTTP (1 mM) at 20 °C.
Table 3.
Rates of Stopped-Flow FRET Changes for Dpo4 Intradomain Mutants
| rate (s−1) |
||||
|---|---|---|---|---|
| domain | mutant | phase | with DNAOH | with DNAH |
| Little Finger | Y274W-R267CCPM | P1 | 6 ± 1 | – |
| P2 | – | – | ||
| Y274W-E291CCPM | P1 | 10 ± 2 | 12 ± 2 | |
| P2 | 0.89 ± 0.04 | – | ||
| Y274W-K329CCPM | P1 | 16 ± 3 | 11 ± 2 | |
| P2 | 0.46 ± 0.02 | – | ||
| Thumb | Y224W-K172CCPM | P1 | 14 ± 5 | – |
| P2 | 0.17 ± 0.04 | – | ||
| Y224W-K212CCPM | P1 | 12 ± 1 | 14 ± 4 | |
| P2 | 0.131 ± 0.008 | – | ||
| Palm | Y118W-K137CCPM | P1 | 9 ± 3 | 5 ± 2 |
| P2 | 0.05 ± 0.02 | – | ||
| Finger | S22W-K56CCPM | P1 | 14 ± 3 | 18 ± 4 |
| P2 | 0.4 ± 0.2 | 0.3 ± 0.1 | ||
| S22W-V62CCPM | P1 | 17 ± 2 | 15 ± 4 | |
| P2 | 0.2 ± 0.1 | 0.2 ± 0.1 | ||
To distinguish between intradomain motions induced by dNTP binding and those associated with the catalysis of nucleotide insertion, we repeated the experiments described above with DNA containing a dideoxy-terminated primer terminus [DNAh (Materials and Methods)]. Interestingly, when phosphodiester bond formation was prohibited, the P2 phase vanished (Figure 5, red traces) and the rates of P1 remained unchanged (Table 3) upon mixing with dTTP for the majority of Dpo4 mutants. Accordingly, P1 must represent a precatalytic conformational change induced by dNTP binding, while the P2 motions must be associated with chemistry or postchemistry events. Consistent with this assignment, the burst phase, single-turnover rates determined in Table 2 for the CPM-labeled Dpo4 mutants agree with the observed rates for P2 at 20 °C (Table 3). However, as previously shown,13 the P2 phase cannot signify the rate-limiting step of nucleotide incorporation but rather signifies the reverse of the conformational changes that developed during nucleotide binding and insertion, including the rate-limiting fine active site adjustments, as well as the global domain conformational transitions. For several of the Dpo4 mutants, the P1 phase could not be well-resolved, likely resulting from the poor signal-to-noise ratios demonstrated by these mutants potentially made worse by reduced nucleotide binding affinities when using the dideoxy-terminated primer.30 Interestingly, the P2 phase for both intradomain Finger mutants (S22W-K56CCPM and S22W-V62CCPM) was still observed (Figure 5E,F), and the phase rates were virtually unaffected (Table 3). These results suggest the presence of additional domain motions following the fast conformational change (P1) despite the dideoxy-terminated primer. Consistently, an interdomain mutant (Y108W-S307CCPM) from an earlier study15 demonstrated a similar P2 phase that was independent of phosphodiester bond formation.
DISCUSSION
It is well-known that proteins are dynamic macromolecules that experience conformational fluctuations in solution despite static crystal structures that would often suggest otherwise.31–37 In fact, the conformational dynamics of DNA polymerases have been extensively studied by us2,13–15,19,38,39 and others,40–44 through a variety of techniques and at each major step of the catalytic cycle to help clarify the enzymatic mechanism of DNA synthesis. Our FRET-based stopped-flow13–15 and single-molecule19 methodologies have allowed the measurement of the kinetic rates of the conformational changes between the distinct structural states (i.e., apo, binary, and ternary) of the model Y-family DNA polymerase, Dpo4.6 Although it is widely accepted that binding of dNTP to the binary complex of DNA polymerases prompts protein conformational changes that substantiate the induced-fit model of nucleotide incorporation and selectivity,20,45–48 the nature of how a polymerase specifically binds to its DNA substrate is less understood.
Usage of both Induced-Fit and Conformational Selection Mechanisms by DNA Polymerases When Binding to DNA
Given the inherent flexibility of macro molecules like polymerases, two mechanisms49–51 exist to describe the binding of a DNA polymerase to DNA: (1) an induced-fit mechanism in which the DNA binds loosely to an inactive form of the polymerase before stimulating conformational changes to an active form and (2) a conformational selection mechanism in which the enzyme is in an equilibrium between inactive and active forms, the latter of which can bind DNA. Our previous stopped-flow FRET15 and computational11 analyses support a complex binding mechanism for binding of Dpo4 to DNA that includes aspects of both the induced-fit and conformational selection mechanisms. Such a combination of binding scenarios has been -suggested for several other biological interactions, as well.50–53 In general, the mechanism involves multiple, interconverting conformational states of an enzyme each with varying binding competency for the substrate, which may bind to the substrate prior to undergoing conformational adjustments that serve to realize the final enzyme•substrate complex. Therefore, while all forms of the enzyme may bind to the substrate, more or fewer conformational adjustments may be necessary to reach the final competent binary complex, depending on the conformational state of the enzyme at the moment of initial substrate encounter. The data collected in this study further support merging of the two binding mechanisms for DNA binding by DNA polymerases.
Expanded DNA Binding Mechanism of Dpo4
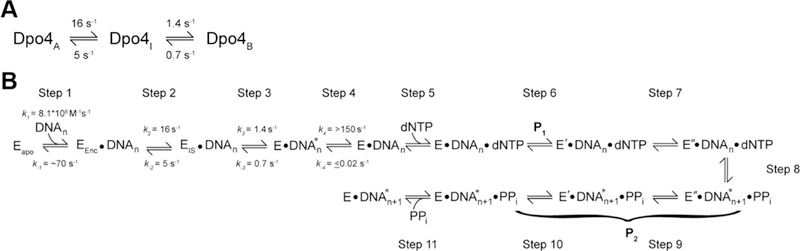
In our previous study,15 a Trp donor in the Palm domain and a CPM acceptor in the LF domain (Y108W-K329CCPM) provided evidence that the conformational change between the apo and DNA-bound states of Dpo4 may occur even in the absence of DNA. These unique findings prompted a follow-up computational study to examine the binding of Dpo4 to DNA.11 It was concluded that the DNA binding mechanism is complex, consisting of a Dpo4 conformational equilibrium among three states (Figure 6A) with different distributions of populations at each binding stage. While the Dpo4A and Dpo4B states refer to those conformations observed in the apo and binary crystal structures, respectively, the Dpo4I state represents an inevitable intermediate that must form during the transition from Dpo4A to Dpo4B. Accordingly, the complete binding process consists of four kinetically connected steps (steps 1–4 in Figure 6B) characterized by their similarity to the final bound state as well as distinct ratios of the Dpo4 conformers. The stopped-flow FRET results presented here provide experimental evidence to validate the computational model and link together previously conflicting reports concerning the DNA binding mechanism of Dpo4. Figure 6B depicts the proposed model for binding of Dpo4 to DNA. The initial bimolecular encounter of Dpo4 with DNA (step 1, Figure 6B) to form the encounter complex (EEnc•DNAn) is nearly diffusion limited (7.8 × 108 M−1 s−1) as estimated in panels A and B of Figure 3. Interestingly, we do not consider it a coincidence that this elusive binding step could be observed through only the LF (Figure 3) and Finger (Figure 4) intradomain FRET systems. It has been previously suggested that the LF domain and the 14-amino acid residue linker joining it to the polymerase core mediate initial binding to DNA through a “fly-casting” mechanism whereby the positively charged domain and linker facilitate the early capture of the DNA through increased flexibility and electrostatic interactions.11,19,54 Furthermore, it is known that a disordered loop in the Finger domain, observed in the apo structure of Dpo4, undergoes a transition to ordered upon DNA binding as shown in the Dpo4 binary complex structure,6 and that this transition is tightly coupled to the degree of DNA binding of the LF domain.11 It is likely that our intradomain FRET system in the Finger domain is sensitive to this transition, resulting in the observed increases in the level of FRET (Figure 4A). Moreover, the lack of appreciable signal changes in the other polymerase domains (Figure S1) upon rapid mixing with DNA supports this assertion. Remarkably, the aforementioned LF domain-facilitated DNA binding and the disordered to ordered transition may be a hallmark of the DNA binding mechanism of the Y-family polymerases.
Figure 6.
Complex DNA binding mechanism of Dpo4. (A) While in the unbound state, Dpo4 is able to interconvert among three conformational states (Dpo4A, Dpo4I, and Dpo4B), each with a different propensity for binding to DNA. Our intradomain analysis has permitted the assignment of forward and reverse rate constants to these transitions. (B) Eapo, EEnc, EIS, E, E′, and E″ represent different conformations of Dpo4 throughout the DNA binding and nucleotide binding and incorporation process. E•DNA* and E•DNA represent nonproductive and productive binary complexes, respectively, connected through a DNA translocation event. P1 and P2 signify the fluorescence phases observed via our FRET technique.
Following the formation of the EEnc•DNAn complex (step 1, Figure 6B), the results of our earlier stopped-flow FRET study15 monitoring the motion between the LF and Palm domains support the inclusion of a succeeding conformational change (step 2, Figure 6B) to form an intermediate complex between Dpo4 and DNA (EIS•DNAn, Figure 6B). During step 2, the LF domain undergoes a transition from its position in the apo state (Dpo4A, Figure 6A), seen in the apo-Dpo4 crystal structure,6 to an intermediate position (Dpo4I, Figure 6A) wherein the complete native contacts of the bound state (Dpo4B, Figure 6A) have not yet fully formed. This is then followed by the rate-limiting step (step 3, Figure 6B) of binding of Dpo4 to DNA as discovered in this study through the slow phase rate (1.5 ± 0.3 s−1) observed in experiments with the intradomain Finger mutant S22W-V62CCPM that was independent of DNA concentration (see Results and Figure 4A). We further validated the inclusion of step 3 (Figure 6B) with an experiment that recorded the time-dependent anisotropy of Alexa488-labeled DNAOH (500 nM) upon rapid mixing with wt Dpo4 (500 nM). Consistently, a slow change in anisotropy was observed (Figure 4B) with a rate (1.2 ± 0.2 s−1) that agreed well with the aforementioned slow phase rate from the experiments with the intradomain Finger mutant, indicating the formation of the E•DNAn* complex (step 3, Figure 6B). During step 3, the polymerase is undergoing the final transition from the intermediate conformation (Dpo4I, Figure 6A) to the bound-state conformation (Dpo4B, Figure 6A) observed in the binary crystal structure.6 The final DNA binding step (step 4, Figure 6B) involves the translocation of the polymerase by one base pair along the DNA (E•DNAn), to vacate the position of the primer–template junction base pair at the active site.13,14 This rapid translocation event (>150 s−1) was demonstrated through the P0 fluorescence phase of our previous stopped-flow FRET analyses.13,14
A previous fluorescence study17 monitoring the emission of a Trp residue (T239W) in the LF domain of Dpo4 reported a rate of dissociation from DNA of ∼70 s−1. Although this dissociation rate is much faster than those determined by 32P-based kinetics7 (0.02 s−1), stopped-flow FRET15 (3.3 s−1), and single-molecule FRET18,19 (0.2–2.5 s−1), their particular fluorescence method used may have been reporting on a specific aspect of the DNA binding mechanism. We suggest that the Trp mutant was ideally suited to monitor the rapid dissociation (step 1, Figure 6B) of the EEnc•DNAn complex (Figure 6B) as the Trp probe was likely very sensitive to the changes in polarity that accompany DNA unbinding in the LF domain. Moreover, the stopped-flow FRET study15 monitoring the conformational motion between the LF and Palm domains could detect the reverse conformational change (5 s−1, step 2, Figure 6B) of Dpo4 from the intermediate state (Dpo4A, Figure 6A) to the apo state (Dpo4A Figure 6A), and fast fluorescence phases observed in the dissociation experiments (Figures 3C,E and 4C) of this study likely report on this event, as well (4.5–9 s−1). Interestingly, the fast dissociation rates measured by a single-molecule study19 of Dpo4 also agree with this assignment (2.5 s−1). The reverse of step 3 is exemplified by the slow fluorescence phases (0.5–1 s−1) observed in the dissociation experiments of this study (Figures 3C–E and 4C,D) which likely represent the slow, reverse isomerization of Dpo4 to unbind the DNA. The slow rate of DNA dissociation (0.02 s−1) measured through the 32P-based kinetic experiments7 must then report on the complete unbinding process from the fully competent bound state (E•DNAn). Taken together, our novel stopped-flow FRET findings and the results of previous studies7,11,13–15,17–19 have uniquely probed different facets of the complex Dpo4–DNA binding process and have collectively permitted the comprehensive elucidation of the complete DNA binding mechanism (Figure 6B).
Subtle Intradomain Motions Collectively Limit Single-Nucleotide Incorporation
For the majority of Dpo4 intradomain mutants, two distinct FRET phases could be observed during nucleotide binding and incorporation. Unfortunately, the distance constraints imposed by our intradomain investigation resulted in suboptimal separations between the FRET donor and acceptor probes for some of the mutants (Table 1), which led to poor signal-to-noise ratios during certain experiments. As such, clear P1 and P2 phases could not be observed for all Dpo4 mutants (i.e., Y274W-R267CCPM and Y224W-K172CCPM) (Figure 5 and Table 3), but this does not necessarily indicate that intradomain motions are not occurring during catalytic events, only that our FRET system is limited in scope.
Our investigation of the conformational motions of individual polymerase domains during nucleotide binding and incorporation is, to the best of our knowledge, the first of its kind. The intradomain FRET changes upon mixing preincubated Dpo4 and DNA with dNTP reveal that subtle motions within each polymerase core domain, as well as the LF domain, occur during nucleotide binding and incorporation (Figure 5 and Table 3). Experiments involving the LF intradomain mutants Y274W-K329CCPM, Y274W-E291CCPM, and Y274W-R267CCPM (Figure 5A–C) expand our earlier discovery15 of a rotational axis in the LF domain perpendicular to helices M and L. Here, we find that in addition to the putative rotation of the entire LF domain reported previously, α-helix L is also dynamic and likely bends to enhance the polymerase grip on the DNA. The Trp donor, Y274W, is located at the distal end of α S-helix L of the LF domain in the relative proximity (10.7 Å, Table 1) of the R267CCPM acceptor residue that is positioned in the middle of the helix (Figure 1). On the basis of the observed fluorescence changes, we suspect that during nucleotide insertion, the end of a-helix L containing the Y274W mutation bends toward the acceptor residues K329CCPM and E291CCPM, and consequently moves away from R267CCPM, in a manner that would seem to strengthen the polymerase–DNA interactions for subsequent nucleotide incorporation. Alternatively, the loops containing K329CCPM and E291CCPM may swing toward the DNA, which would lead to a similar FRET change. Comparison of multiple, ternary complex crystal structures of Dpo4 bound to undamaged DNA,4 or abasic site-containing DNA,5 and dNTp5 reveals that the mobile LF domain likely allows proper DNA substrate alignment and orientation for catalysis. This observation has allowed for the idea that Dpo4 executes its role as a lesion bypass polymerase through an “induced-grip” approach wherein the LF domain modulates primer–template positioning within the active site to facilitate translesion synthesis.55 Our intradomain FRET system has thus provided a glimpse of the LF domain dynamics that are applied during nucleotide binding to enhance protein–DNA interactions for efficient catalysis on undamaged DNA. As previously shown for the global conformational motions of Dpo4,14 we expect that the intradomain motions shown here will be altered by a DNA lesion.
Apart from Y274W-R267CCPM and Y224W-K172CCPM, the remaining intradomain mutants displayed characteristic P1 and P2 transitions during correct nucleotide insertion, and the P2 phase was not observed when using a dideoxy-terminated primer for all mutants (Figure 5 and Table 3), except for those of the Finger domain, suggesting that slow intradomain motions occur following nucleotide incorporation as we rationalized previously.13 For the Finger intradomain mutants, the P2 transition occurred in the same direction as P1 and was independent of phosphodiester bond formation (Figure 5E,F). The reason is unclear, and we speculate that the observed slow intradomain motions in the Finger domain reflect subtle conformational transitions following the initial binding of the correct nucleotide that function to align active site residues around the replicating base pair.4,5 As expected, this alignment occurs even when catalysis is prohibited by a chain-terminating primer (DNAH, Figure 5E,F, red trace). Interestingly, the rate of this conformational motion (∼0.3 s−1, Table 3) is on the order of the rate-limiting step of single-nucleotide incorporation (kburst, Table 2) as measured by 32P-based kinetic assays.7,9,15 Furthermore, it is likely that the dynamic motions observed within each polymerase domain during nucleotide incorporation (P2) collectively contribute to active site rearrangements (step 7, Figure 6B), which we hypothesize to be the rate-determining step of single-nucleotide incorporation.13 In fact, subtle adjustments of loops and other secondary structural elements within each domain of Dpo4 between the binary and ternary complexes have been observed structurally,6 as well as distinct changes in rotamer configurations of amino acid side chains in the Palm (Y10) and Finger (Y48) domain that reside in the nucleotide binding pocket of Dpo4.
While the results presented here, and those of our previous stopped-flow FRET studies,13–15 have provided significant insight into the solution-state behavior of Dpo4 during substrate binding and catalysis, higher-resolution, solution-state methodologies are necessary to fully evaluate the conformational changes that take place throughout the reaction pathway. Advancements in macromolecular nuclear magnetic resonance56 will permit the atomic-level inspection of Dpo4 motions at a multitude of time scales, and the assignments of the backbone nitrogen, carbon, and amide proton resonances of the polymerase core57 and LF domain58 will strongly inform these future investigations.
Taken together, the results presented here elucidate the complex DNA binding mechanism of a Y-family DNA polymerase that involves aspects of both induced-fit and conformational selection mechanisms. Furthermore, intradomain protein motions were observed throughout nucleotide binding and incorporation, some of which may kinetically limit the rate of correct nucleotide incorporation. Future investigations of the effects of proliferating cell nuclear antigen (PCNA) and DNA damage on the intradomain motions of Dpo4 are of particular intrigue.
Supplementary Material
ACKNOWLEDGMENTS
We thank Andrew Reed and Walter Zahurancik for their careful editing of the manuscript.
Funding
This work was supported by both National Institutes of Health Grant R21ES024585 and National Science Foundation Grant MCB-0960961 to Z.S. A.T.R. was supported by National Institutes of Health Training Grant T32GM008512.
ABBREVIATIONS
- Dpo4
DNA polymerase IV of S. solfataricus
- FRET
Förster resonance energy transfer
- LF
Little Finger
- CPM
7-(diethylamino)-3-(4′-maleimidylphenyl)-4-methylcoumarin
- DNAOH
normal primer–template DNA substrate
- DNAH
dideoxy-terminated primer–template DNA substrate; wt, wild type
- wt
wild type
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bio-chem.6b00878.
Figures showing stopped-flow traces of several Dpo4 mutants binding to DNA and unbinding from DNA and stopped-flow control experiments (PDF)
REFERENCES
- (1).Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, and Woodgate R (2001) The Y-family of DNA polymerases. Mol. Cell 8, 7–8. [DOI] [PubMed] [Google Scholar]
- (2).Maxwell BA, and Suo Z (2014) Recent insight into the kinetic mechanisms and conformational dynamics of Y-Family DNA polymerases. Biochemistry 53, 2804–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Fowler JD, and Suo Z (2006) Biochemical, structural, and physiological characterization of terminal deoxynucleotidyl transferase. Chem. Rev 106, 2092–2110. [DOI] [PubMed] [Google Scholar]
- (4).Ling H, Boudsocq F, Woodgate R, and Yang W (2001) Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell 107, 91–102. [DOI] [PubMed] [Google Scholar]
- (5).Ling H, Boudsocq F, Woodgate R, and Yang W (2004) Snapshots of replication through an abasic lesion; structural basis for base substitutions and frameshifts. Mol. Cell 13, 751–762. [DOI] [PubMed] [Google Scholar]
- (6).Wong JH, Fiala KA, Suo Z, and Ling H (2008) Snapshots of a Y-family DNA polymerase in replication: substrate-induced conformational transitions and implications for fidelity of Dpo4. J. Mol. Biol 379, 317–330. [DOI] [PubMed] [Google Scholar]
- (7).Fiala KA, and Suo Z (2004) Mechanism of DNA polymerization catalyzed by Sulfolobus solfataricus P2 DNA polymerase IV. Biochemistry 43, 2116–2125. [DOI] [PubMed] [Google Scholar]
- (8).Fiala KA, and Suo Z (2004) Pre-steady-state kinetic studies of the fidelity of Sulfolobus solfataricus P2 DNA polymerase IV. Biochemistry 43, 2106–2115. [DOI] [PubMed] [Google Scholar]
- (9).Fiala KA, Sherrer SM, Brown JA, and Suo Z (2008) Mechanistic consequences of temperature on DNA polymerization catalyzed by a Y-family DNA polymerase. Nucleic Acids Res. 36, 1990–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Maxwell BA, and Suo Z (2012) Kinetic basis for the differing response to an oxidative lesion by a replicative and a lesion bypass DNA polymerase from Sulfolobus solfataricus. Biochemistry 51, 3485–3496. [DOI] [PubMed] [Google Scholar]
- (11).Chu X, Liu F, Maxwell BA, Wang Y, Suo Z, Wang H, Han W, and Wang J (2014) Dynamic conformational change regulates the protein-DNA recognition: an investigation on binding of a Y-family polymerase to its target DNA. PLoS Comput. Biol 10, e1003804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Boudsocq F, Kokoska RJ, Plosky BS, Vaisman A, Ling H, Kunkel TA, Yang W, and Woodgate R (2004) Investigating the role of the little finger domain of Y-family DNA polymerases in low fidelity synthesis and translesion replication. J. Biol. Chem 279, 32932–32940. [DOI] [PubMed] [Google Scholar]
- (13).Xu C, Maxwell BA, Brown JA, Zhang L, and Suo Z (2009) Global conformational dynamics of a Y-family DNA polymerase during catalysis. PLoS Biol. 7, e1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Maxwell BA, Xu C, and Suo Z (2012) DNA lesion alters global conformational dynamics of Y-family DNA polymerase during catalysis. J. Biol. Chem 287, 13040–13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Maxwell BA, Xu C, and Suo Z (2014) Conformational dynamics of a Y-family DNA polymerase during substrate binding and catalysis as revealed by interdomain Forster resonance energy transfer. Biochemistry 53, 1768–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Eoff RL, Sanchez-Ponce R, and Guengerich FP (2009) Conformational changes during nucleotide selection by Sulfolobus solfataricus DNA polymerase Dpo4. J. Biol. Chem 284, 21090–21099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Beckman JW, Wang Q, and Guengerich FP (2008) Kinetic analysis of correct nucleotide insertion by a Y-family DNA polymerase reveals conformational changes both prior to and following phosphodiester bond formation as detected by tryptophan fluorescence. J. Biol. Chem 283, 36711–36723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Brenlla A, Markiewicz RP, Rueda D, and Romano LJ (2014) Nucleotide selection by the Y-family DNA polymerase Dpo4 involves template translocation and misalignment. Nucleic Acids Res. 42, 2555–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Raper AT, Gadkari VV, Maxwell BA, and Suo Z (2016) Single-Molecule Investigation of Response to Oxidative DNA Damage by a Y-Family DNA Polymerase. Biochemistry 55, 2187–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Joyce CM, and Benkovic SJ (2004) DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry 43, 14317–14324. [DOI] [PubMed] [Google Scholar]
- (21).Doublie S, Sawaya MR, and Ellenberger T (1999) An open and closed case for all polymerases. Structure 7, R31–35. [DOI] [PubMed] [Google Scholar]
- (22).Wong I, Patel SS, and Johnson KA (1991) An induced-fit kinetic mechanism for DNA replication fidelity: direct measurement by single-turnover kinetics. Biochemistry 30, 526–537. [DOI] [PubMed] [Google Scholar]
- (23).Clegg RM (1992) Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol 211, 353–388. [DOI] [PubMed] [Google Scholar]
- (24).Lakowicz JR (2006) Principles of Fluorescence Spectroscopy, pp 954, Springer, Berlin. [Google Scholar]
- (25).Johnson KA (1992) 1 Transient-State Kinetic Analysis of Enzyme Reaction Pathways 20, 1–61. [Google Scholar]
- (26).Alley SC, Shier VK, Abel-Santos E, Sexton DJ, Soumillion P, and Benkovic SJ (1999) Sliding clamp of the bacteriophage T4 polymerase has open and closed subunit interfaces in solution. Biochemistry 38, 7696–7709. [DOI] [PubMed] [Google Scholar]
- (27).Trakselis MA, Alley SC, Abel-Santos E, and Benkovic SJ (2001) Creating a dynamic picture of the sliding clamp during T4 DNA polymerase holoenzyme assembly by using fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U. S. A 98, 8368–8375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Chakraborty S, Ittah V, Bai P, Luo L, Haas E, and Peng Z (2001) Structure and dynamics of the alpha-lactalbumin molten globule: fluorescence studies using proteins containing a single tryptophan residue. Biochemistry 40, 7228–7238. [DOI] [PubMed] [Google Scholar]
- (29).Zhou G, Wong MT, and Zhou GQ (1983) Diffusioncontrolled reactions of enzymes. An approximate analytic solution of Chou’s model. Biophys. Chem 18, 125–132. [DOI] [PubMed] [Google Scholar]
- (30).Astatke M, Grindley ND, and Joyce CM (1998) How E. coli DNA polymerase I (Klenow fragment) distinguishes between deoxy- and dideoxynucleotides. J. Mol. Biol 278, 147–165. [DOI] [PubMed] [Google Scholar]
- (31).Henzler-Wildman K, and Kern D (2007) Dynamic personalities of proteins. Nature 450, 964–972. [DOI] [PubMed] [Google Scholar]
- (32).Benkovic SJ, and Hammes-Schiffer S (2003) A perspective on enzyme catalysis. Science 301, 1196–1202. [DOI] [PubMed] [Google Scholar]
- (33).Koshland DE (1958) Application of a Theory of Enzyme Specificity to Protein Synthesis. Proc. Natl. Acad. Sci. U. S. A 44, 98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Kumar S, Ma B, Tsai CJ, Sinha N, and Nussinov R (2000) Folding and binding cascades: dynamic landscapes and population shifts. Protein Sci. 9, 10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Tsai CJ, Ma B, Sham YY, Kumar S, and Nussinov R (2001) Structured disorder and conformational selection. Proteins: Struct., Funct., Genet 44, 418–427. [DOI] [PubMed] [Google Scholar]
- (36).Bosshard HR (2001) Molecular recognition by induced fit: how fit is the concept? News Physiol. Sci 16, 171–173. [DOI] [PubMed] [Google Scholar]
- (37).Okazaki K, and Takada S (2008) Dynamic energy landscape view of coupled binding and protein conformational change: induced-fit versus population-shift mechanisms. Proc. Natl. Acad. Sci. U. S. A 105, 11182–11187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Xu C, Maxwell BA, and Suo Z (2014) Conformational dynamics of Thermus aquaticus DNA polymerase I during catalysis. J. Mol. Biol 426, 2901–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Maxwell BA, and Suo Z (2013) Single-molecule investigation of substrate binding kinetics and protein conformational dynamics of a B-family replicative DNA polymerase. J. Biol. Chem 288, 11590–11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).DeLucia AM, Grindley ND, and Joyce CM (2007) Conformational changes during normal and error-prone incorporation of nucleotides by a Y-family DNA polymerase detected by 2-aminopurine fluorescence. Biochemistry 46, 10790–10803. [DOI] [PubMed] [Google Scholar]
- (41).Zhang H, Cao W, Zakharova E, Konigsberg W, and De La Cruz EM (2007) Fluorescence of 2-aminopurine reveals rapid conformational changes in the RB69 DNA polymerase-primer/ template complexes upon binding and incorporation of matched deoxynucleoside triphosphates. Nucleic Acids Res. 35, 6052–6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Purohit V, Grindley ND, and Joyce CM (2003) Use of 2-aminopurine fluorescence to examine conformational changes during nucleotide incorporation by DNA polymerase I (Klenow fragment). Biochemistry 42, 10200–10211. [DOI] [PubMed] [Google Scholar]
- (43).Rothwell PJ, Mitaksov V, and Waksman G (2005) Motions of the fingers subdomain of klentaq1 are fast and not rate limiting: implications for the molecular basis of fidelity in DNA polymerases. Mol. Cell 19, 345–355. [DOI] [PubMed] [Google Scholar]
- (44).Federley RG, and Romano LJ (2010) DNA polymerase: structural homology, conformational dynamics, and the effects of carcinogenic DNA adducts. J. Nucleic Acids 2010, 457176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Tsai YC, and Johnson KA (2006) A new paradigm for DNA polymerase specificity. Biochemistry 45, 9675–9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Post CB, and Ray WJ Jr. (1995) Reexamination of induced fit as a determinant of substrate specificity in enzymatic reactions. Biochemistry 34, 15881–15885. [DOI] [PubMed] [Google Scholar]
- (47).Johnson KA (1993) Conformational coupling in DNA polymerase fidelity. Annu. Rev. Biochem 62, 685–713. [DOI] [PubMed] [Google Scholar]
- (48).Rothwell PJ, and Waksman G (2005) Structure and mechanism of DNA polymerases. Adv. Protein Chem 71, 401–440. [DOI] [PubMed] [Google Scholar]
- (49).Zhou HX (2010) From induced fit to conformational selection: a continuum of binding mechanism controlled by the timescale of conformational transitions. Biophys. J 98, L15–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Hammes GG, Chang YC, and Oas TG (2009) Conformational selection or induced fit: a flux description of reaction mechanism. Proc. Natl. Acad. Sci. U. S. A 106, 13737–13741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Csermely P, Palotai R, and Nussinov R (2010) Induced fit, conformational selection and independent dynamic segments: an extended view of binding events. Trends Biochem. Sci 35, 539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Wlodarski T, and Zagrovic B (2009) Conformational selection and induced fit mechanism underlie specificity in noncovalent interactions with ubiquitin. Proc. Natl. Acad. Sci. U. S. A 106, 19346–19351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Grunberg R, Leckner J, and Nilges M (2004) Complementarity of structure ensembles in protein-protein binding. Structure 12, 2125–2136. [DOI] [PubMed] [Google Scholar]
- (54).Shoemaker BA, Portman JJ, and Wolynes PG (2000) Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc. Natl. Acad. Sci. U. S. A 97, 8868–8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Fleck O, and Schar P (2004) Translesion DNA synthesis: little fingers teach tolerance. Curr. Biol 14, R389–391. [DOI] [PubMed] [Google Scholar]
- (56).Kleckner IR, and Foster MP (2011) An introduction to NMR-based approaches for measuring protein dynamics. Biochim. Biophys. Acta, Proteins Proteomics 1814, 942–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Ma D, Fowler JD, Yuan C, and Suo Z (2010) Backbone assignment of the catalytic core of a Y-family DNA polymerase. Biomol. NMR Assignments 4, 207–209. [DOI] [PubMed] [Google Scholar]
- (58).Ma D, Fowler JD, and Suo Z (2011) Backbone assignment of the little finger domain of a Y-family DNA polymerase. Biomol. NMR Assignments 5, 195–198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.