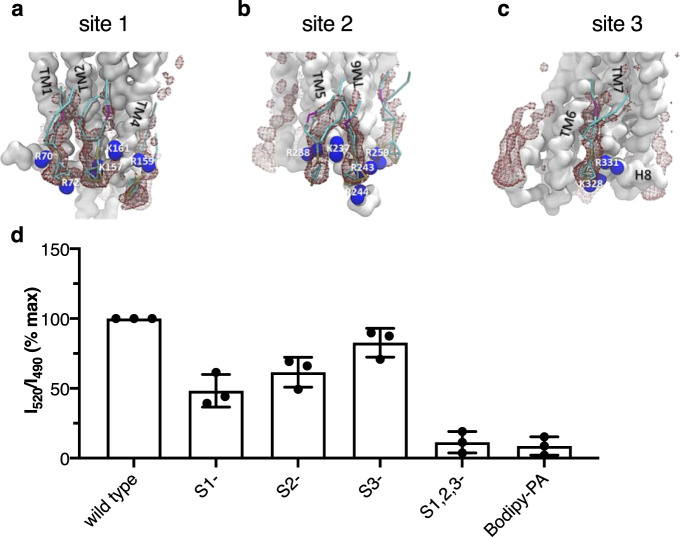
Fig. 2. PIP2-binding sites.
a–c Representative snapshots showing the position of the lipid(s) in the three sites (active conformation) that were most occupied by PIP2 (a site 1; b site 2; c site 3). The residues interacting with the lipid within each of these sites are indicated (blue), the PIP2 molecules are reported in sticks colored according to the CG beads and the receptor backbone (active conformation) as a white surface. d FRET signal between Bodipy-FL PIP2 (2.5 molar%) and Lumi-4 Tb attached to C2556.27 of the wild-type receptor or of the putative PIP2-binding sites mutants. The FRET signal obtained with negative control labeled lipid Bodipy-PA is given for comparison. The signal was normalized to that measured with wild-type GHSR. Data in (d) are mean ± SD of three experiments and is provided as a Source data file.