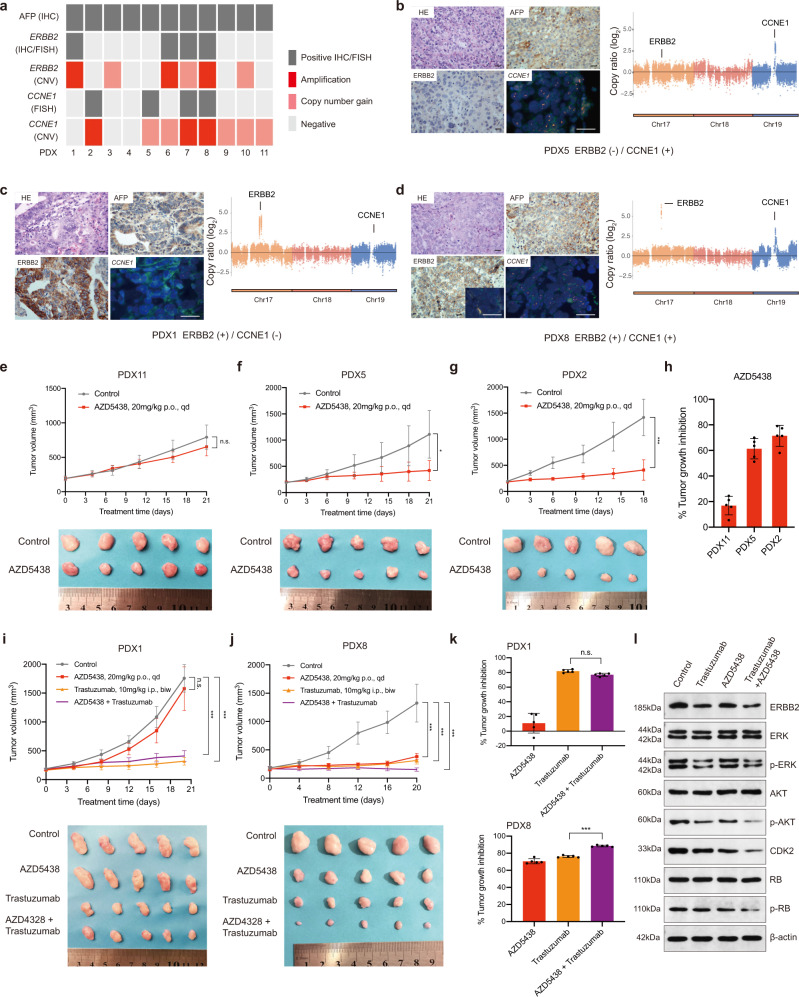
Fig. 5. Validation of selected genes as potential therapeutic targets in PDX models of AFPGC.
a The status of AFP, ERBB2, and CCNE1 in AFPGC PDX models. The CNVs status of ERBB2 and CCNE1 are defined by the WES results of corresponding patients. b–d The representatives of AFPGC PDX models with different status of ERBB2 and CCNE1. Scale bar represents 50 μm. Data were obtained from three independent experiments. The right panel represents the CNVs status of ERBB2 and CCNE1 in the corresponding patients. e–h In vivo sensitivity of AFPGC with or without CCNE1 amplification to CCNE1 inhibition (AZD5438) (n = 5 biologically independent samples). Tumor volumes and proportion of tumor growth inhibition were shown as means ± SD. i–k In vivo sensitivity of ERBB2-positive AFPGC with or without CCNE1 amplification to ERBB2 (Trastuzumab) and CCNE1 (AZD5438) dual inhibition (n = 5 biologically independent samples). Tumor volumes and proportion of tumor growth inhibition were shown as means ± SD. l The western blot analysis of critical molecules (ERBB2, ERK, p-ERK, AKT, p-AKT, CDK2, RB, and p-RB) for ERBB2 and CCNE1 co-positive PDX models (PDX8) were performed. The western blotting images are representative of three different independent experiments. Statistical significance was determined using two-sided Student’s t-test (e–g) and one-way ANOVA (i–k). biw, twice a week; Chr, chromosome; CNVs, copy number variations; FISH, fluorescence in situ hybridization; HE, hematoxylin–eosin staining; IHC, immunohistochemistry; i.p, intraperitoneal injection; kD, kilodaltion; p.o, oral gavage; qd, per day. *P < 0.05, **P < 0.01, ***P < 0.001; n.s., not significant. Source data are provided as a Source Data file.