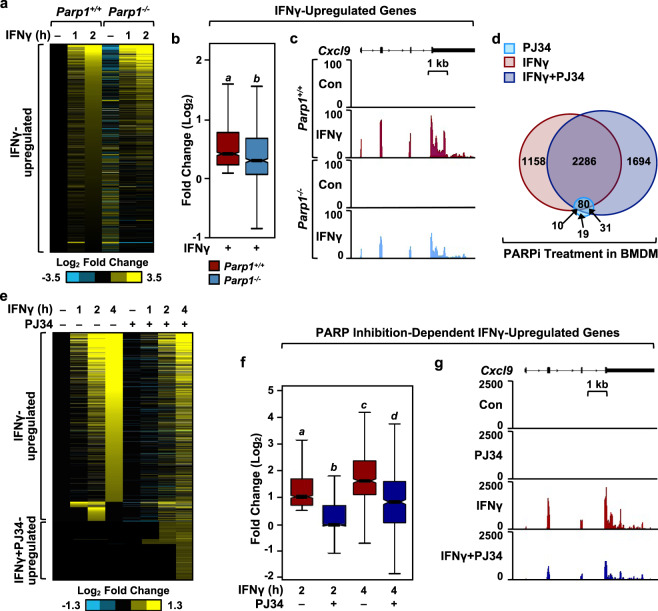
Fig. 1. PARP-1 regulates IFNγ-dependent gene expression in bone marrow-derived macrophages (BMDMs).
a Heatmap of RNA-seq data representing changes in the expression of IFNγ-regulated genes from mRNA-seq in BMDMs from wild-type (Parp1+/+) or Parp1 knockout (Parp1-/-) mice. The cells were treated with IFNγ for the indicated times. b, c Box plots (b) and browser tracks (c) illustrating IFNγ-stimulated gene expression in Parp1+/+ or Parp1-/- mice. BMDM cells were treated with IFNγ for 2 h and steady-state mRNA levels from RNA-seq were expressed as fold change relative to the untreated control. Boxes represent 25th–75th percentile (line at median) with whiskers at 1.5*IQR. Boxes marked with different letters are significantly different from each other (Wilcoxon Signed-Rank test; p < 2.2 × 10−16). Box plots represent 960 genes. d Venn diagrams showing differentially regulated genes from RNA-seq in BMDMs upon treatment with PJ34 (light blue), IFNγ (red), or IFNγ + PJ34 (blue). Numbers indicate the number of differentially regulated genes compared to the untreated control. e Heatmap of RNA-seq data representing the changes in gene expression of IFNγ- regulated genes upon co-treatment with PJ34. f, g PARP-1 catalytic activity is required for IFNγ-dependent gene expression in BMDMs. Box plots (f) and browser tracks (g) representing changes in gene expression from RNA-seq upon IFNγ treatment ± PJ34 (n = 1053 genes; Wilcoxon Signed-Rank test; p < 2.2 × 10−16). Boxes represent 25th–75th percentile (line at median) with whiskers at 1.5*IQR.