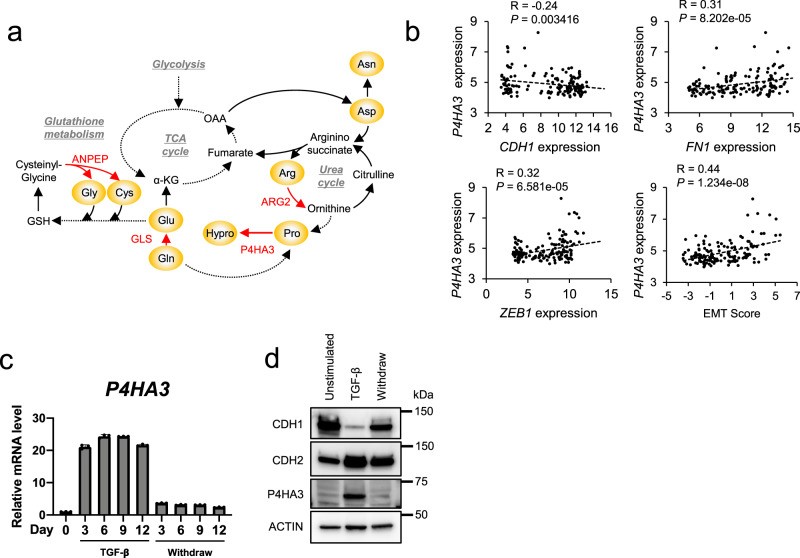
Fig. 4. Identification of P4HA3 by integrated metabolomic and transcriptomic analysis.
a Metabolic reactions of ARG2, ANPEP, GLS, and P4HA3. b Correlations between EMT markers and P4HA3 mRNA expression in non-small cell lung cancer (NSCLC) cell lines in the Cancer Cell Line Encyclopedia (CCLE) dataset. EMT scores were calculated based on the expression of the reported 76 genes as an EMT marker in NSCLC. c Expression of P4HA3 during TGF-β-stimulation and withdrawal. A549 cells were stimulated with TGFβ (2 ng/mL) for the indicated number of days (marked TGF-β), and mRNA level of P4HA3 was measured by real-time PCR. Values are presented as the mean ± SD from triplicate samples. d Protein levels of EMT markers (CDH1 and CDH2) and P4HA3 in A549 cells. A549 cells were stimulated with TGF-β (5 ng/mL) for 3 days (marked TGF-β) and then cultured in normal growth medium without TGF-β for 3 days (marked Withdraw). Actin was used as a loading control for western blot analysis.