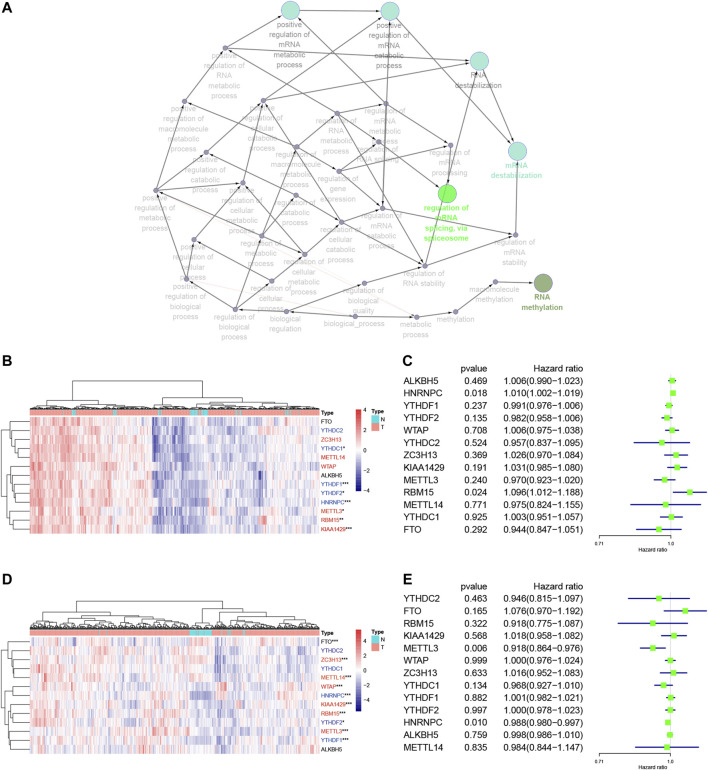
FIGURE 2.
Association of m6A RNA methylation regulatory genes with NSCLC prognosis. (A) The GO terms and KEGG enrichment pathway analysis of the m6A regulator gene, the different colors represent the different pathways. (B) Differential expression of these 13 m6A RNA methylated regulator genes in LUAD (red: “writer”; blue: “readers”; black: “erasers”). (C) Forest plot of the univariate Cox regression analytic data. The 13 m6A RNA methylation regulators in LUAD were analyzed using the univariate Cox regression and the data are plotted using the forest plot. (D) Differential expression of these 13 m6A RNA methylated regulators in LUSC (red: “writer”; blue: “readers”; black: “erasers”). (E) Forest plot of the univariate Cox regression analysis. The 13 m6A RNA methylation regulators in LUAD were analyzed using the univariate Cox regression and the data are plotted using the forest plot. ***p < 0.001, **p < 0.01, and *p < 0.05. The KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, Gene Ontology; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; m6A, N6-methyladenosine; N, Normal; T, Tumor.