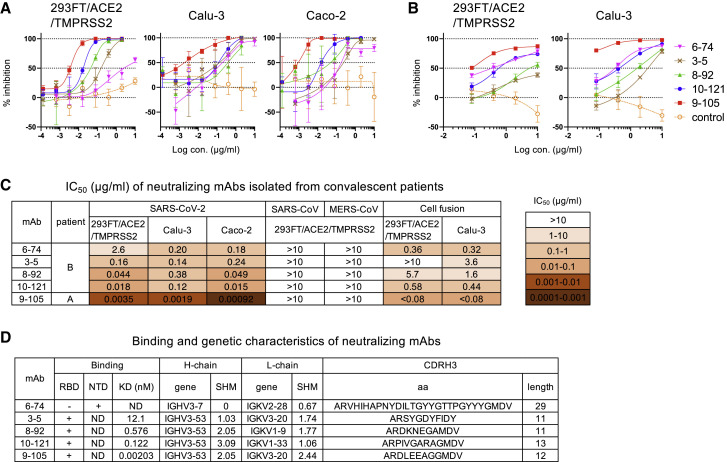
Figure 2.
Characterization of five neutralizing mAbs
(A) Neutralization of SARS-CoV-2 pseudovirus by mAbs 6-74, 3-5, 8-92, 10-121, and 9-105 in 293FT/ACE2/TMPRSS2, Calu-3 (human lung cancer cell line), and Caco-2 (human colon adenocarcinoma cell line) cells is shown. A non-neutralizing mAb, 8-38, was used as a negative control (orange open circle).
(B) Cell fusion of 293FT/DSP8-11/SARS-CoV-2-S cells with 293FT/DSP1-7/ACE2/TMPRSS2 and Calu-3/DSP1-7 cells was measured by luciferase activity at 6 and 20 h after coculture, respectively. A non-neutralizing mAb, 5-76, was used as a negative control (orange open circle).
(C) IC50 values of the five mAbs are summarized.
(D) The binding and genetic characteristics of mAbs are summarized. Binding to RBD and NTD was analyzed by AlphaScreen. KD values were determined by SPR analysis (Figure S1D). Gene usage (gene), somatic hypermutation % (SHM) of heavy and light chains, and CDRH3 amino acids (aa) and length were analyzed by IMGT vquest.
Data shown in (A) and (B) are represented as means ± SD (n = 3). See also Figure S1.