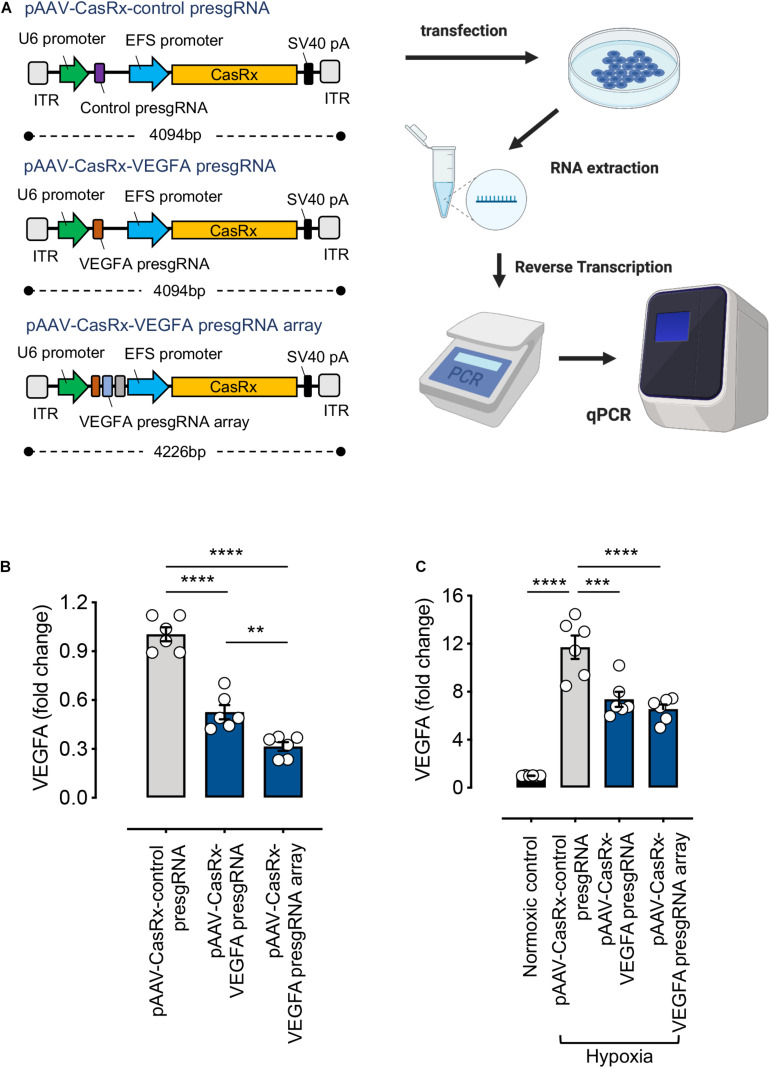
FIGURE 3.
RNA-editing efficiency of the CRISPR/CasRx system using the all-in-one pAAV construct. (A) Illustration of the all-in-one pAAV constructs and the experimental procedure. Created with BioRender.com. (B) HEK293FT cells were transfected with pAAV–CasRx–control pre-sgRNA, pAAV–CasRx–VEGFA pre-sgRNA, and pAAV–CasRx–VEGFA pre-sgRNA array for 72 h. The VEGFA mRNA level was determined by quantitative PCR (qPCR) as described in section “METHODS” (n = 6). (C) Human Müller cells (MIO-M1) were transfected with pAAV–CasRx–control pre-sgRNA, pAAV–CasRx–VEGFA pre-sgRNA, and pAAV–CasRx–VEGFA pre-sgRNA array for 48 h and under hypoxia condition for another 24 h. The VEGFA mRNA level was determined by qPCR as described in section “METHODS” (n = 6). Data are expressed as the mean ± SEM. Statistical analysis was performed with GraphPad Prism 7 (GraphPad, San Diego, CA, United States) and undertaken with one-way ANOVA and Tukey’s multiple comparison test. **P < 0.01, ***P < 0.001, ****P < 0.0001.