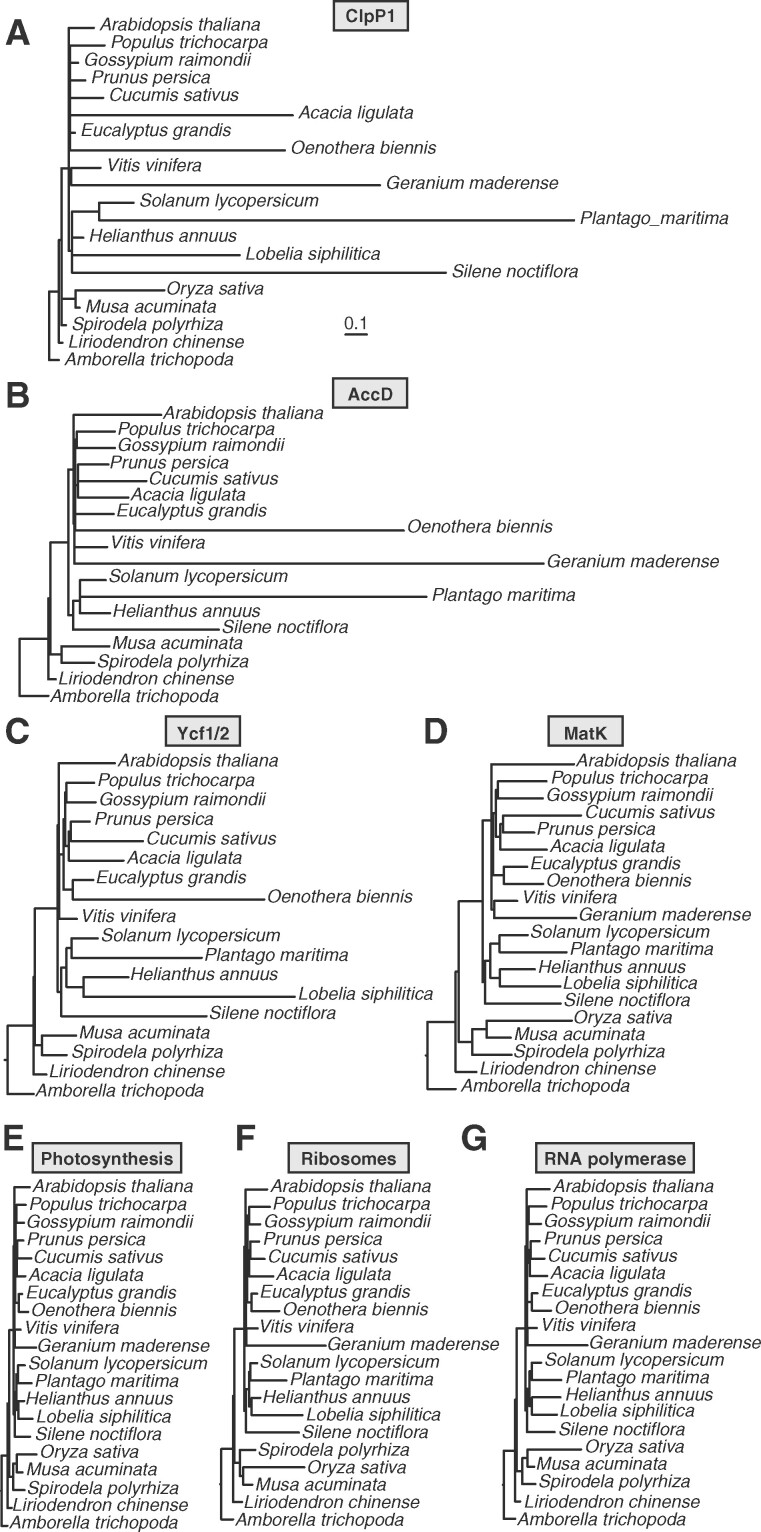
Figure 1.
Trees based on plastome partitions. Branch length-optimized trees inferred from amino acid sequence alignments for plastid genes partitioned into seven functional categories (as described in Supplemental Table 2). Branch lengths are shown on the same scale for all trees to highlight differences in rates of amino acid evolution between partitions. Each plastome partition tree was used for ERC analysis against all nuclear gene trees.