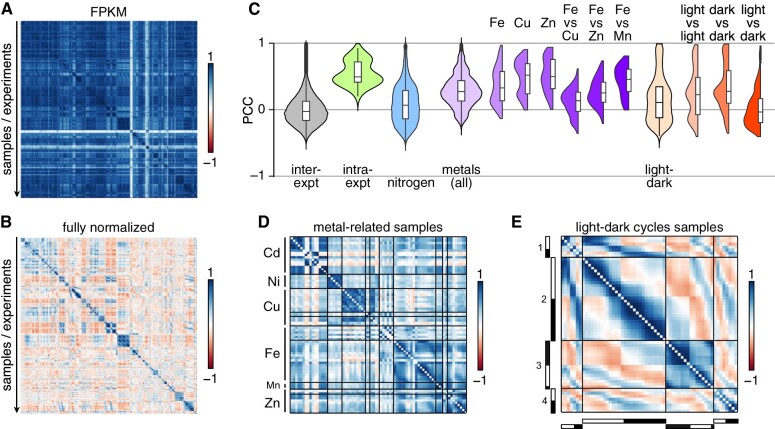
Figure 1.
Samples from the same experiment are strongly correlated. A), Correlation matrices between all samples using expression estimates for all 17,741 nuclear genes as FPKM. B), As in panel A, but after all normalization steps. In panels A and B, samples belonging to the same experiment are in consecutive order, and roughly in chronological order. C), Distribution of PCCs between (inter-expt, gray) and within (intra-expt, green) experiments. PCCs for all comparisons between experiments are shown as violin plots and box plots, alongside mean PCCs from all samples within each experiment, samples collected in the context of nitrogen deprivation (blue), PCCs for all metal-related samples (light purple) and specific metals (darker shades of purple), samples collected over a diurnal cycle (light orange), and PCC between subsets of samples (darker shades of orange). Values along the diagonal of the matrix (equal to 1) were discarded prior to plotting. D), Correlation matrix for samples from metal-related experiments, all from the Merchant laboratory, and in which either one micronutrient has been omitted from the growth medium (for deficiency conditions: copper Cu, iron Fe, manganese Mn, and zinc Zn) or a toxic metal was added to observe the effect on homeostasis (cadmium Cd and nickel Ni). E), Correlation matrix of samples collected over a diurnal cycle. The light- and dark-part of each sampling day is indicated on the left and bottom sides of the matrix as white and black bars, respectively. Four time courses are compared here (Panchy et al., 2014; Zones et al., 2015; Strenkert et al., 2019).