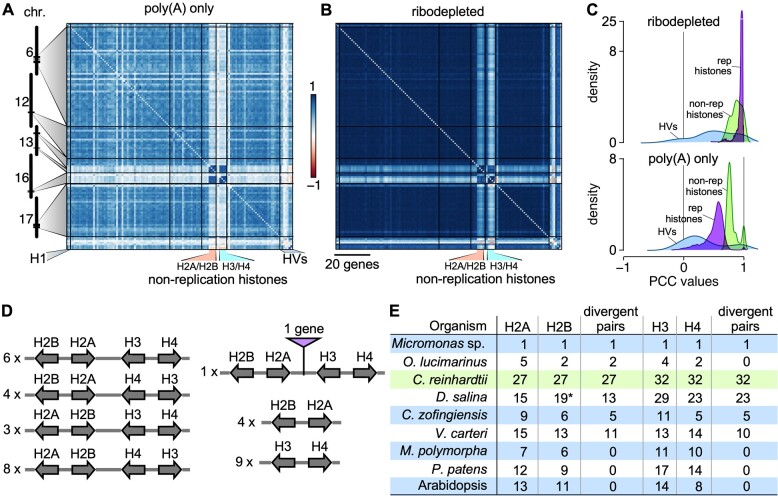
Figure 5.
Correlations between Chlamydomonas histone genes. A), Correlation matrix among Chlamydomonas histone genes, ordered according to their genomic coordinates, using RNA-seq data derived from poly(A)-selected samples. B), Same as (A), using RNA-seq data derived from ribodepleted samples. Histone genes that are not regulated by the cell cycle are indicated as “non-replication histones.” H1, histone H1 genes; HVs, histone variants. C), Distribution of PCCs for classes of histones genes shown in (A) and (B). Histone variants (HVs) are shown in light blue, replication-associated histones in purple, and non-replication histones in light green. D), Global clustering of histone genes in Chlamydomonas. All histone genes occur as divergent pairs and are oftentimes grouped as one representative of each major histone type (H2A, H2B, H3, and H4). The number to the left gives the number of instances of the given arrangement in the Chlamydomonas genome. E), Comparison of histone gene clustering in selected photosynthetic organisms. O. lucimarinus, Ostreococcus lucimarinus; D. salina, Dunaliella salina; V. carteri, Volvox carteri; C. zofingiensis, Chromochloris zofingiensis; M. polymorpha, Marchantia polymorpha; P. patens, Physcomitrium patens. The asterisk for Histone H2B genes in D. salina indicates that they are absent from the current annotation, but were identified by TBLASTN against the D. salina genome with Chlamydomonas histone H2B protein sequence as query.