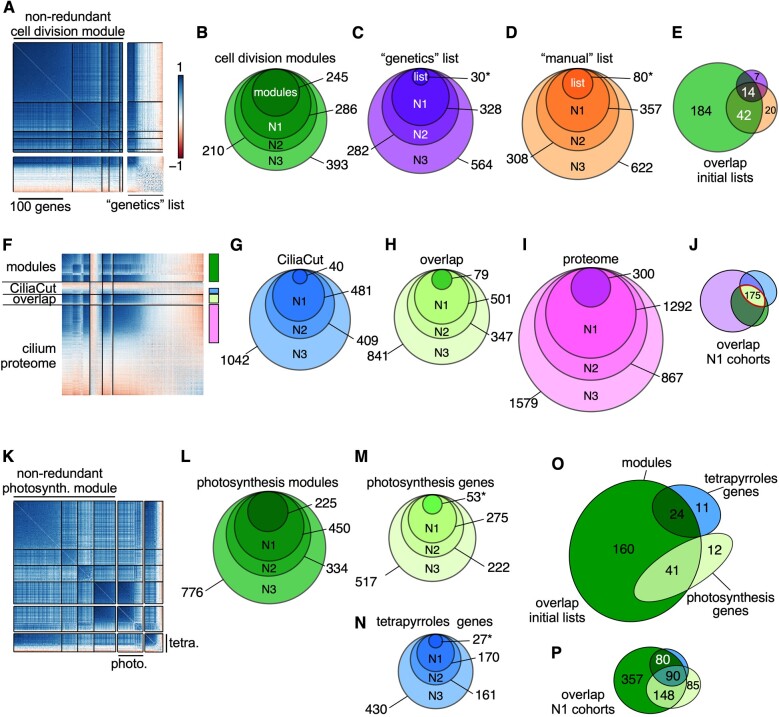
Figure 6.
Core cell division genes are coordinately and highly co-expressed. A), Correlation matrix of non-redundant cell division modules and correlation matrix of genes whose loss of function leads to cell division defects (Tulin and Cross, 2014; Breker et al., 2018). Genes within each set were ordered according to hierarchical clustering using the FPC method in corrplot. B–D), Co-expressed cohorts, shown as nested Venn diagrams, associated with genes from the cell division modules (B), the genetics list (C), or genes involved in DNA replication and chromosome segregation (manual list) (D) from networks N1–N3. E), Overlap between original gene lists related to cell division (modules, genetics, and manual lists). F), Correlation matrix of non-redundant cilia modules (modules) and genes belonging to CiliaCut only (CiliaCut), the cilium proteome and shared genes between CiliaCut and the cilium proteome (overlap). The color bars on the right refer to the color scheme used for co-expression cohorts in G–J. G–I), Co-expressed cohorts, shown as nested Venn diagrams, associated with genes from CiliaCut (G), the overlap between CiliaCut and the cilium proteome (H), and the cilium proteome (I) from networks N1–N3. J), Overlap between N1 cohorts associated with each initial gene list (CiliaCut, overlap, and cilium proteome). K), Correlation matrix of non-redundant photosynthesis modules, photosynthesis-related genes, and tetrapyrrole biosynthesis-related genes. L–N), Co-expressed cohorts, shown as nested Venn diagrams, associated with genes from the photosynthesis modules (L), photosynthesis-related genes (M), and tetrapyrrole biosynthesis-related genes (N) from networks N1–N3. O), Overlap between initial gene lists. P), Overlap between N1 cohorts associated with photosynthesis and tetrapyrrole biosynthesis. In panels C, D, M, and N, the asterisk indicates that the gene list was restricted to highly co-expressed genes, based on FPC clustering of the data.