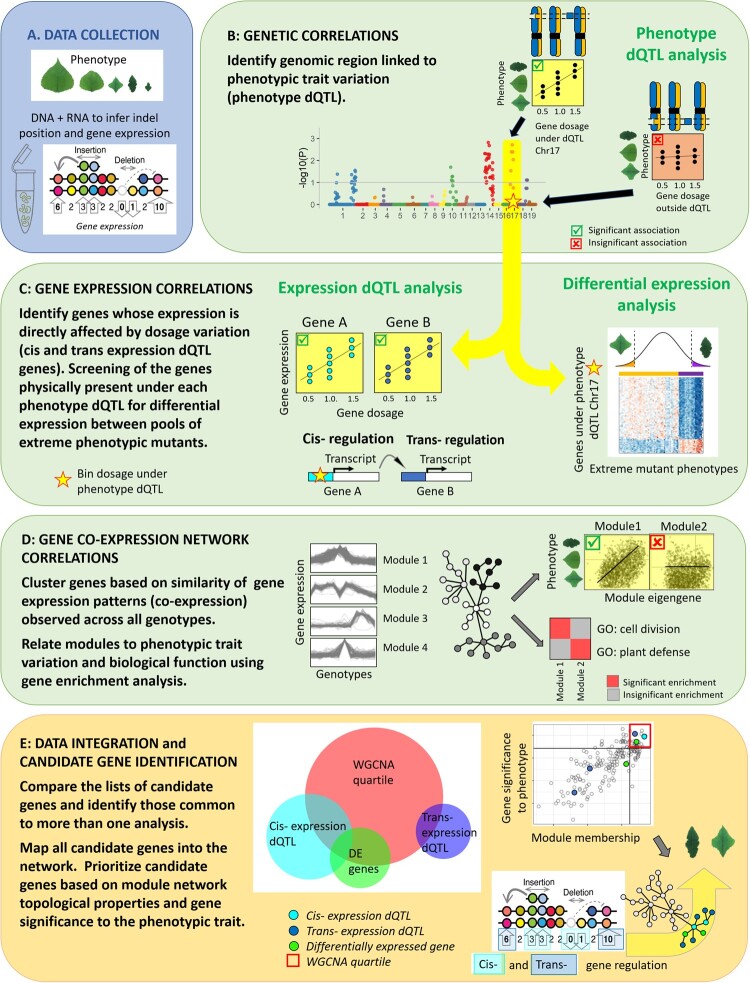
Figure 1.
Overview of the analytic workflow used to investigate the genomic architecture of indel mutation on individual gene expression and co-expression networks. (A) Populus indel lines were characterized for various leaf morphometric traits, genomically characterized for indel positions, and assayed for gene expression in leaf tissues. (B) Genetic correlations were used to identify chromosomal bins in which gene dosage variation was proportional to the phenotypic values (phenotype dQTL). (C) Gene expression correlations were used to identify the genes for which expression was proportional to gene dosage at individual phenotype dQTL bins (expression dQTL). Distinction was made between local (cis-), and distant (trans-) regulation based on the physical position of the gene relative to the chromosomal bin evaluated. In addition, a differential expression analysis between extreme phenotypic mutants was performed to identify candidate genes within the phenotype dQTL bins. (D) Co-expression network analysis: modules of genes that were co-expressed across the various lines were identified. Such modules were then related to the phenotypic traits by evaluating the level of correlation of the module eigengene expression with trait variation, and to biological function using a gene enrichment analysis. (E) All data were integrated to reveal the genomic architecture of the regions and genes regulating leaf morphometric traits. The interactions that are at play between expression dQTL and differentially expressed genes, and their importance toward the phenotype and global biochemical pathways they encode, was further evaluated in the broader context of the gene co-expression network. Candidate genes were mapped onto the network modules, and further evaluated for their importance as key regulator of the module (module membership) and gene significance to the phenotypic trait.