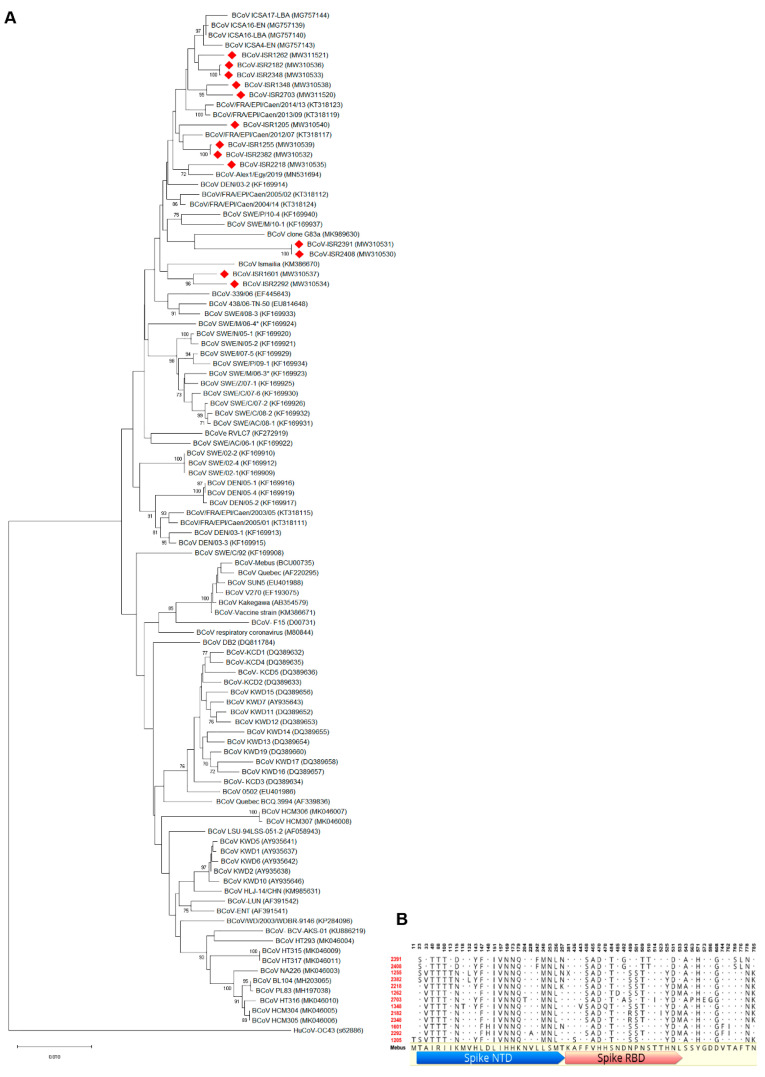
Figure 2.
Phylogenetic tree of the partial Spike gene of Bovine coronavirus (BCoV) from local isolates. (A) Phylogenetic tree of S1 region of BCoV spike glycoprotein. BCoV genomic RNA was isolated from feces of WD diseased cattle and used for molecular characterization of the S1 region. Local isolates (labeled in red) were aligned with selected reference strains. Phylogenetic tree generated based on S1 nucleotide sequences (nucleotides 1–2731) was generated via the neighbor-joining method with bootstrap analysis (1000 replicates, >70%). The scale bar shows the number of substitutions per site. We used HCoV OC43 as an outgroup strain. Spike partial sequences were retrieved from GenBank and embedded in the figure for each strain. (B) Deduced amino acid sequences of S1 from local isolates aligned to reference strain Mebus. Local isolate sequences were aligned using Geneious protein alignment tool to show only disagreements residues compared to the reference sequence of Mebus strain.