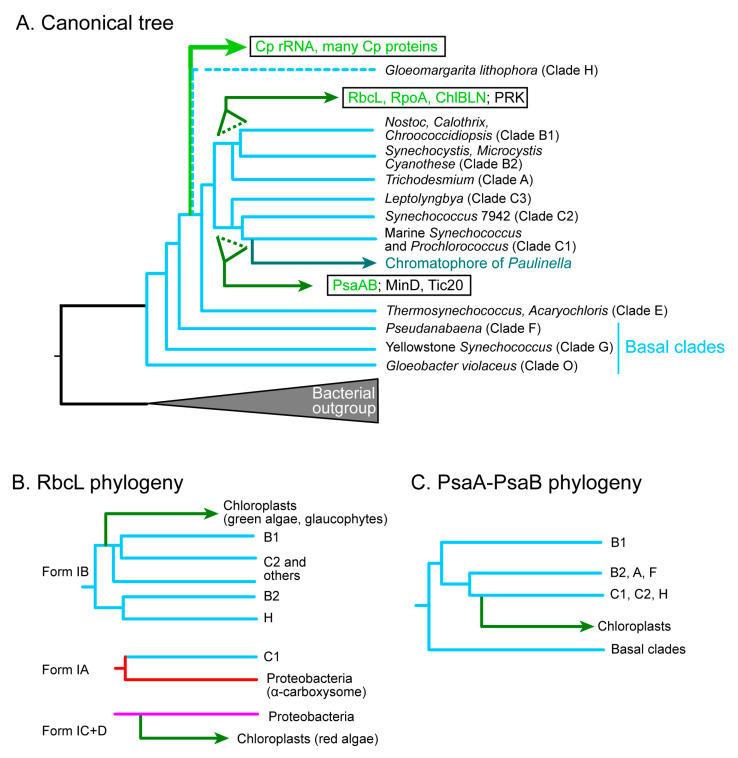
Figure 3.
Schematic phylogenetic trees showing different origins of chloroplast enzymes (or RNA) within the cyanobacterial diversity. (A) Canonical phylogenetic tree of cyanobacteria and chloroplasts. Various clades of cyanobacteria are shown according to [53,54]. Chloroplast-encoded proteins and RNA are shown in green. Nuclear-encoded chloroplast proteins are shown in black. The clade names are taken from [53]. According to [54], G. lithophora is sister to the main chloroplast clade represented by chloroplast rRNA. For simplicity, Clade H is used for G. lithophora. The positions of branching of RbcL and PsaA/B are shown with dotted lines because they do not match exactly with the canonical tree. (B) Different origins of chloroplast and cyanobacterial RbcL. (C) Probable origin of PsaA and PsaB. As explained in the text and Supplementary Materials, the phylogeny of PsaA and PsaB is very difficult. This diagram, which is consistent with previous results, is still a plausible hypothesis among many other alternatives. Different line colors are used to show different lineages such as cyan for cyanobacteria and green for chloroplast. Colored protein names are used to show chloroplast-encoded proteins.