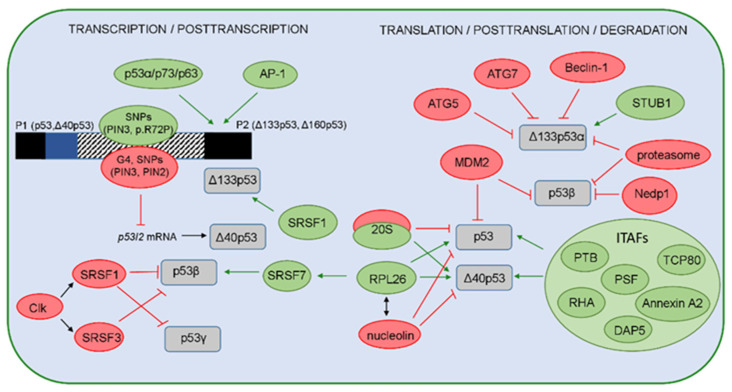
Figure 3.
A model representing regulation of the p53 isoforms’ expression and stability. The positive regulators are shown in green, while the negative regulators are shown in red. On the transcriptional level, expression of the p53 isoforms is regulated by usage of two different promoters P1 or P2, producing the long (p53, Δ40p53) or the short (Δ133p53, Δ160p53) isoforms, respectively. Several regulators can influence the P2 activity. The canonical p53 and p53 family members (p63 and p73 and their isoforms) are shown to transactivate P2 with different efficiency. The transcription from P2 can be activated by the AP-1 transcription factor that mediates the expression of Δ133p53 in H. pylori-infected cells. The single-nucleotide polymorphisms (SNP) and their haplotypes in the internal promoter region (shown as box with a striped pattern) that comprises intron 3, exon 4, and intron 4 can affect the P2 activities. Furthermore, the G4 structures, PIN3 and PIN2 polymorphisms, can decrease the level of the p53I2 mRNA that encodes the Δ40p53 isoforms. The p53 isoforms are regulated on the posttranscriptional level by different splicing factors. SRSF1 and SRSF3, activated by Clk, promote complete exclusion of intron 9 and, thus, negatively regulate the level of p53β and p53γ isoforms. However, SRSF1 upregulates the Δ133p53α expression in human aortic smooth muscle cells. In addition, the binding of RPL26 to the TP53 pre-mRNA allows the recruitment of SRSF7 that prompts alternative splicing and, thus, generates p53β isoforms. Due to IRES, the level of p53 and Δ40p53 is regulated by ITAFs (PTB, Annexin A2, PSF, DAP5, TCP80, RHA) or proteins such as RPL26 or nucleolin. Interestingly, Δ40p53 can be generated by the 20S proteasome that degrades the full-length p53 protein. The level of the full-length p53 protein is regulated by MDM2 that was shown to promote the degradation of p53β. In addition, the level of p53β is also regulated by the MDM2-dependent neddylation, proteasome, and deneddylating enzyme Nedp1. The level of the Δ133p53α isoform is regulated by the proteasome, as well as via autophagic degradation, upon replicative senescence, where the proautophagic proteins (ATG5, ATG7, Beclin-1) act as positive regulators, while the STUB1/CHIP acts as a negative regulator of Δ133p53α degradation and senescence.