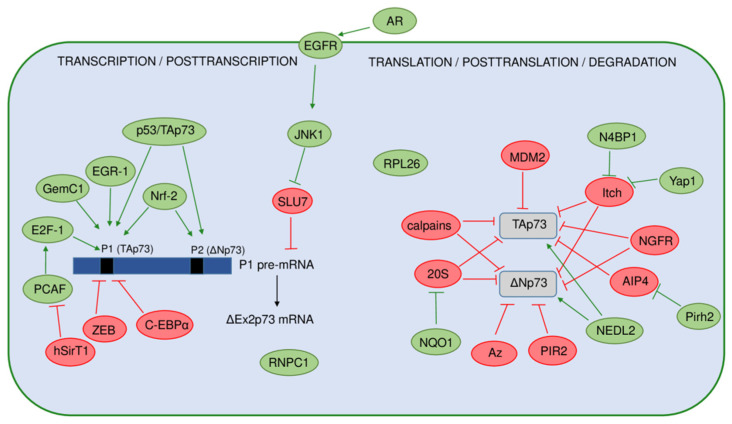
Figure 4.
A model representing regulation of the p73 isoforms’ expression and stability. The positive regulators are shown in green, and the negative regulators are shown in red. On the transcriptional level, expression of the p73 isoforms is regulated by usage of two different promoters (P1 and P2) producing the TAp73 or ΔNp73 isoforms. Black bars represent the TP73 gene promoters. Several regulators specifically activate the P1 promoter, with transcription factor E2F-1 being the most important. In contrast, transcription from P1 is repressed by ZEB, C-EBPα, and hSirT1. The full-length p53 and the TAp73 proteins, as well as Nrf-2, have been found to induce both promoters. On the posttranscriptional level, the alternative splicing of P1 pre-mRNA, leading to increased expression of the ΔEx2p73 isoform, is induced by the activation of EGFR by its ligand amphiregulin (AR) in hepatocellular carcinoma cells. The expression of the ΔEx2p73 isoform is enabled by the inhibition of the RNA splicing factor SLU7 through JNK1 signaling. The stability of the p73 mRNA is increased by the RNA-binding protein RNPC1. Ribosomal protein RPL26 has been found to regulate the p73 translation and protein stability. The p73 protein isoforms are also extensively regulated on a posttranslational level. Here are shown different regulators of the TAp73 and ΔNp73 isoforms’ stability and degradation. Some of them target both TAp73 and ΔNp73 isoforms including Itch, NGFR, calpains, and NQO1 (through 20S proteasome). On the contrary, antizyme (Az) pathway and ligase PIR2 specifically target the ΔNp73 isoforms for degradation after DNA damage.