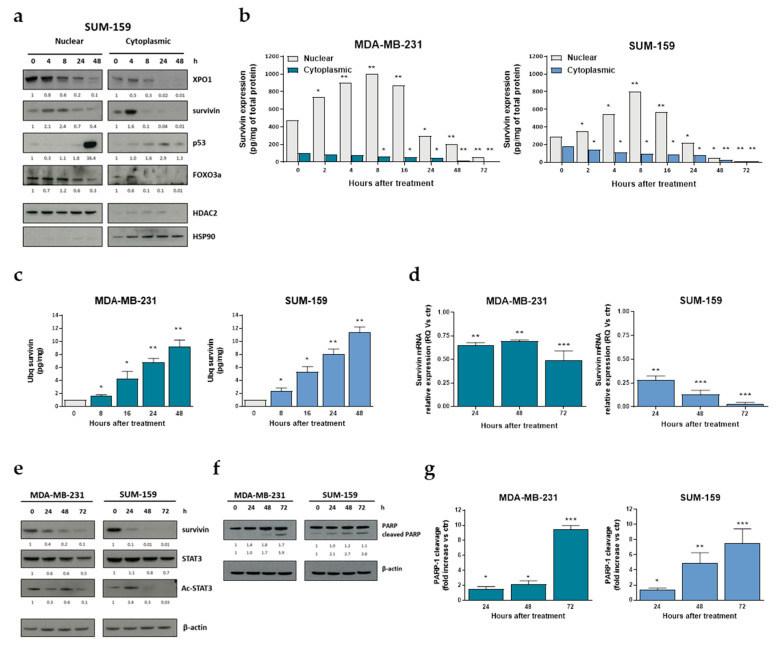
Figure 2.
Effect of selinexor on survivin expression in TNBC cell lines. (a) Representative western blot showing nuclear and cytoplasmic fractions of XPO1 and survivin in TNBC cells exposed to selinexor (IC50 at 72 h, as reported in Table 1). HDAC2 and HSP90 were used to confirm equal protein loading on the gel and to show the relative purity of the nuclear and cytosolic fractions, respectively. Cropped images of selected proteins are shown. For each protein, band density was quantified using ImageJ normalized to loading control and referred to respective untreated control.(b) Quantification of nuclear and cytosolic survivin protein levels by ELISA assay in TNBC cells exposed to selinexor (IC50 at 72 h). Data are reported as amount (pg) of survivin normalized to total (mg) protein extract, and represent the mean values ± SD of three independent experiments. (c) Quantification of ubiquitinated (Ubq)-nuclear survivin protein levels by ELISA assay in TNBC cells exposed to selinexor (IC50 at 72 h). Data are reported as amount (pg) of Ubq-nuclear survivin normalized to total (mg) nuclear protein extract, and represent the mean values ± SD of three independent experiments. (d) Quantification of survivin mRNA expression levels by qRT-PCR in TNBC cells exposed to selinexor (IC50 at 72 h) at different intervals. Data are reported as relative quantity (RQ) in selinexor-treated cells with respect to control cells exposed to 0.01% DMSO (ctr), and represent the mean values ± SD of three independent experiments. (e) Representative western blotting showing the expression of survivin, STAT3 and Ac-STAT3 in TNBC cells exposed to selinexor (IC50 at 72 h) at different intervals. β-actin was used to confirm equal protein loading on the gel. Cropped images of selected proteins are shown. (f) Representative western blotting showing the cleavage of PARP-1 in selinexor-treated (IC50 at 72 h) TNBC cell lines. Cropped images of selected proteins are shown. (g) Quantification of PARP-1 cleavage. Data are expressed as mean values ± SD of three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.005, Student′s t-test.