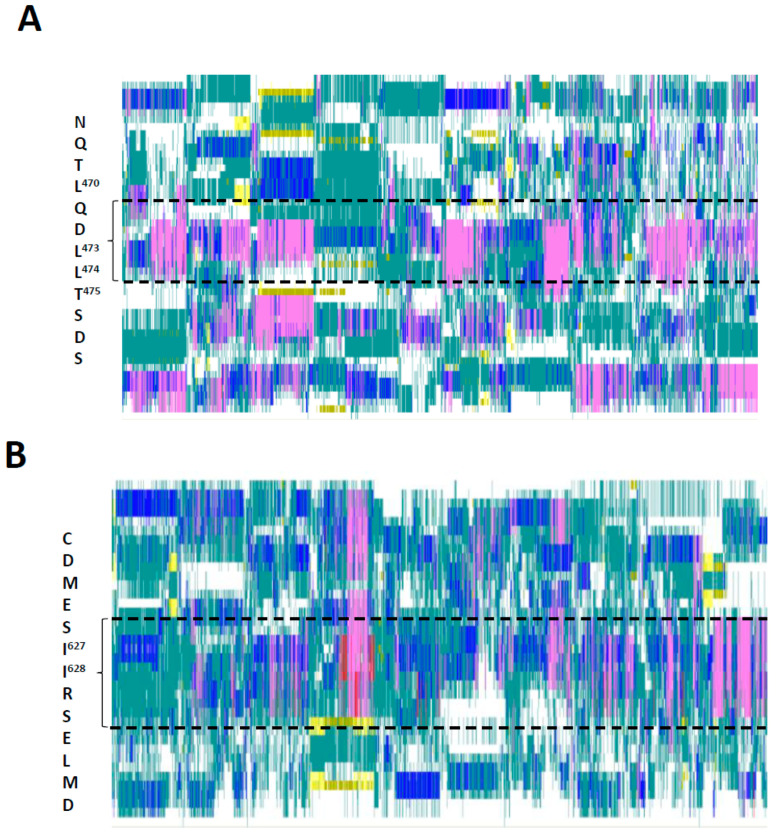
Figure 7.
Secondary structures of the KIX-Binding Region and Transactivation Domain (TAD) formed in ten implicit solvation simulations (gb8_md#1—gb8_md#10), each representing 500 nanoseconds. The positions and sequences of α-helical regions shown are based on experimentally determined locations of KIX-binding motifs [17]. (A) The region (residues 451 to 500) surrounding the α-helix participating in binding to the KIX-domain of the CBP/p300 coactivator complex is shown. The position and sequence of the interaction helix (FOXO467−478) binding to the KIX domain directly is highlighted. (B) The region (residues 606 to 644) surrounding another α-helix participating in binding to the KIX-domain of the CBP/p300 coactivator complex is shown. The position and sequence of the interaction helix (FOXO622−634) binding to the KIX domain directly is highlighted. The color codes represent the secondary structures formed in each simulation frame at each position of the primary amino acid sequence (pink, α-helix; dark blue, π-helix; red, 310 helix; yellow, extended conformation; cyan, turn; white, coil). Data visualized with VMD [34].