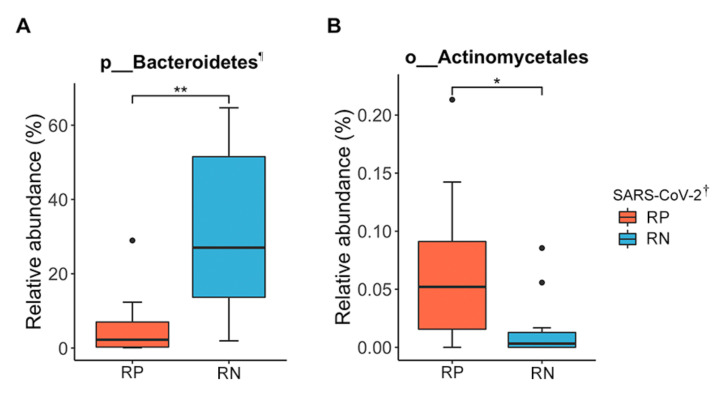
Figure 3.
Significantly different taxa profiles of gut microbiota between infected (respiratory positive (RP)-SARS-CoV-2) and recovered (respiratory negative (RN)-SARS-CoV-2) states of patients with COVID-19. (A) The relative abundance of phylum Bacteroidetes. ¶ The phylum Bacteroidetes, class Bacteroidia, and order Bacteroidales showed the same abundance and statistically significant differences. (B) The relative abundance of the order Actinomycetales. The RP-SARS-CoV-2 as a baseline, the two time points as a fixed effect, and subjects’ IDs as a random effect were used in the mixed model using MaAsLin2. * FDR q < 0.05, ** FDR q < 0.01. † SARS-CoV-2 RNA from the respiratory tract. RP, respiratory positive SARS-CoV-2; RN, respiratory negative SARS-CoV-2.