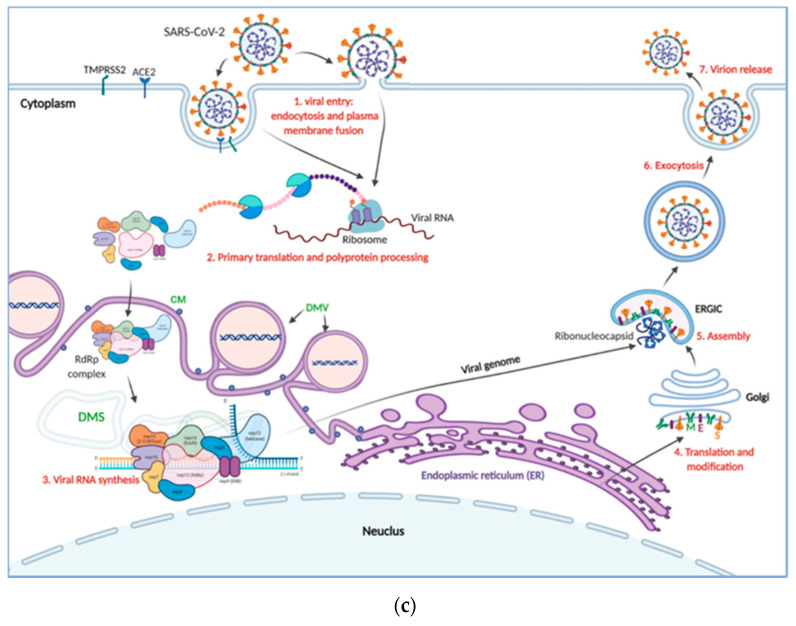
Figure 1.
SARS-CoV-2 genome organization and replication cycle. (a) The SARS-CoV-2 virion structure. While the viral membrane and envelope proteins ensure its genomic RNA incorporation and assembly, the trimeric spike (S) protein provides specificity and high-affinity binding for its receptor to enter into target cells. The positive-sense, single-stranded RNA genome (+ssRNA) is encapsidated by the nucleocapsid. (b) Schematic depiction of SARS-CoV-2 genome architecture and the poly-(proteins) it encodes. (c) Schematics of SARS-CoV-2 replication: (1) SARS-CoV-2 virions bind to ACE2, its cellular receptor, and the type 2 transmembrane serine protease (TMPRSS2), a host factor that promotes viral particles entry and fusion at the plasma membrane or endosomes. (2) In the cytosol, the incoming gRNA will be released and subjected to immediate translation of the ORF1a and ORF1b open reading frame resulting in the polyproteins (pp1a and pp1ab), which are further proteolytically processed to the components of the replicase complex. (3) The replicase complex is an assemblage of multiple nonstructural proteins (nsps) that orchestrate transcription of the viral RNAs and viral replication. Concordantly, the virus compels the host cell to create the viral replication organelles, including the perinuclear double-membrane vesicles (DMV), the convoluted membranes, and small, open double-membrane spherules (DMS). These structures provide a protective microenvironment for the viral gRNA and subgenomic mRNA during replication and transcription. (4,5) The newly synthesized surface structural proteins translocate to the ER membrane and transit through the ER-to-Golgi intermediate compartment (ERGIC), which then assembles and encapsidates the ribonucleocapsid. (6,7) Ultimately, the progeny virions bud into the lumen of the secretory vesicles and will be released by exocytosis.