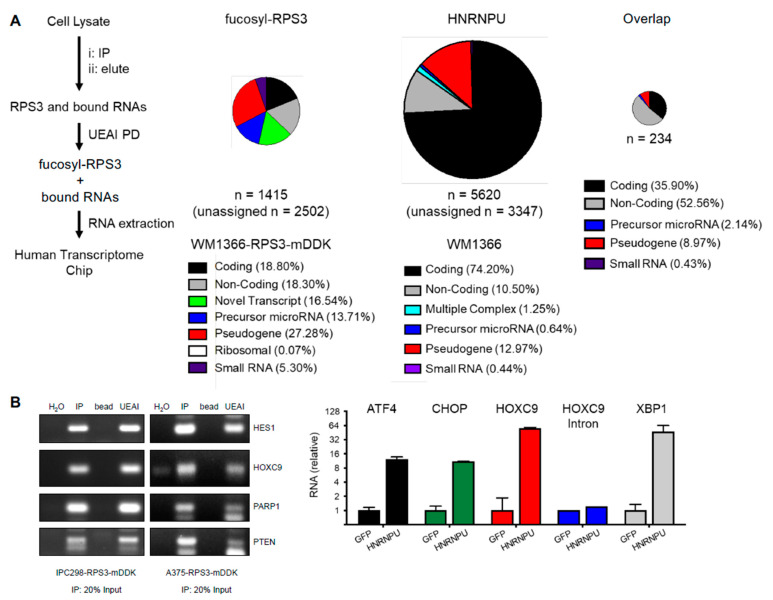
Figure 4.
Identification of RNAs that interact with fucosylated RPS3 and HNRNPU. (A) Workflow and results for fucosylated RPS3 (fucosyl-RPS3) and HNRNPU RIP-Chip. RNAs isolated by fucosylated RPS3 PD, HNRNPU IP, or IgG control IP were analyzed on a transcriptome-level expression array chip. RNAs detected at > 2-fold higher in fucosylated RPS3 PD or HNRNPU IP relative to IgG control IP were counted as a specific interaction. Results were analyzed to assign RNAs to one of several distinct subclasses of RNA. (B) The RNAs identified in the RIP-Chip were validated by RIP-PCR and RIP-qPCR. Coding RNAs for HES1, HOXC9, PARP1, and PTEN were validated by PCR for the fucosylated RPS3 RIP (left). Coding RNAs for ATF4, CHOP, HOXC9, and XBP1 were validated by qPCR for interaction with HNRNPU. HOXC9 intron was tested to determine whether HNRNPU interacts with mature RNAs, as opposed to unprocessed, pre-spliced transcripts.