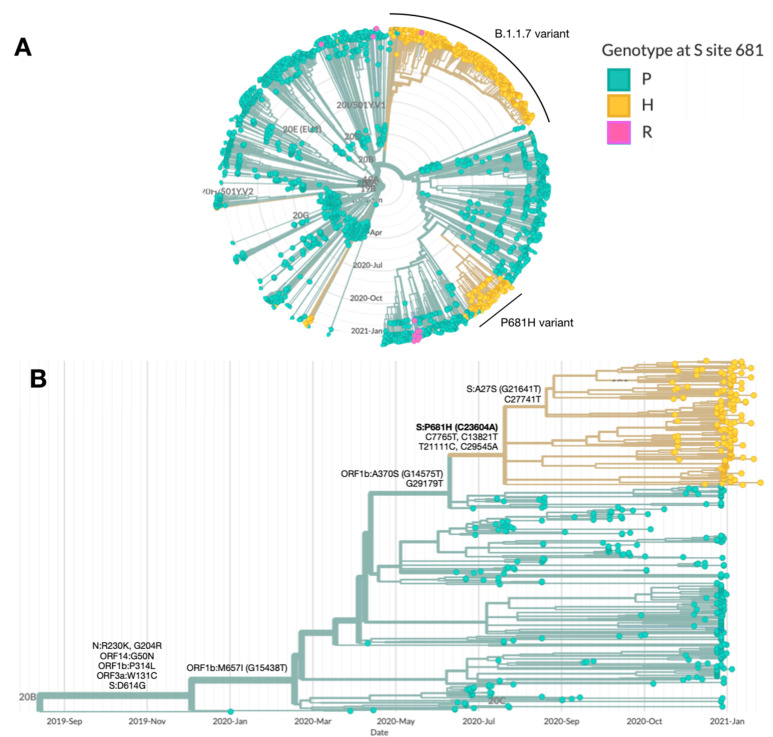
Figure 1.
Characterization of the B.1.1.50 + P681H variant. Phylogenetic trees highlighting the genotype at site 681 in the S protein, with the wild-type proline (P) in green and the mutations histidine (H) and arginine (R) in yellow and blue, respectively. (A) SARS-CoV-2 genomes sequenced in Israel from March 2020 to January 2021 (n = 2482). The main clusters harboring the P681H mutation (in yellow) are the B.1.1.7 (20I/501Y.V1) and the B.1.1.50 + P681H variant. (B) SARS-CoV-2 genomes from of the B.1.1.50 lineage only (n = 489). The B.1.1.50 + P681H cluster is composed of local (Israel) viruses only, with the exception of 2 sequences from isolates identified in the Palestinian authorities, and includes a sub-cluster with an additional S protein non-synonymous mutation, A27S. Additional mutations are listed by each branch. Phylogenetic trees were created with Nextstrain Augur pipeline and visualized with Auspice [3]. Non-Israeli B.1.1.50 lineage sequences were downloaded from GISAID and identified with Pangolin classification [4].