Figure 3.
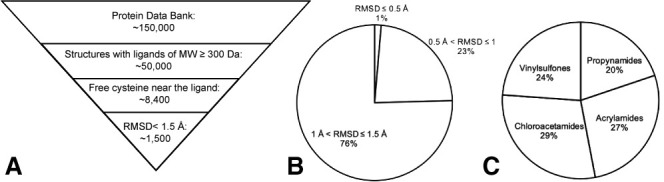
PDB-wide application of covalentizer identifies candidate irreversible inhibitors for more than 1,500 structures
(A) We filtered the PDB for structures that had only protein chains (no DNA/RNA), and contained a small molecule of at least 300 Da. This threshold was set to ensure some minimal initial fit/binding affinity to the target, as well as to filter out non-ligand small molecules, such as crystallization reagents. We used a PyMOL-based script to filter only the structures in which at least one ligand atom is <6 Å away from the sulfur atom of a cysteine residue. This cysteine also has to be free (no disulfide or other covalent modifications). After running the covalentizer protocol and filtering only for results with <1.5 Å RMSD of the MCS between the reversible ligand and the covalent analog generated by covalentizer, there were 1,553 structures for which at least one such prediction was obtained.
(B) The top 1% of results have an RMSD under 0.5 Å; 23% are between 0.5 and 1 Å, and 76% are between 1 and 1.5 Å.
(C) The distribution of the four electrophiles used is balanced, with 29% chloroacetamides, 27% acrylamides, 24% vinylsulfones, and 20% propynamides.
