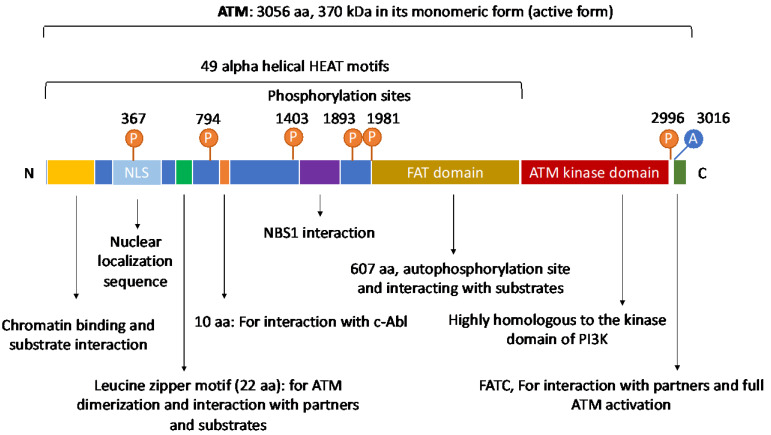
Figure 1.
Human ATM’s domain map. ATM is a large gene with 66 exons and consists of multiple domains that allow its protein to directly interact with numerous regulators, partners, and downstream targets. In its N terminus, ATM contains a Nuclear Localization Signal that enables its nuclear translocation. ATM also uses its N-terminus for interacting with chromatin, substrates and partners, for instance, p53, LKB1, NBS1, among others. The putative leucine zipper motif on ATM N-terminus is documented to be essential for its dimerization and interaction with other partners or substrates. On its C-terminus, ATM has a FAT (FRAP, ATM and TRRAP proteins) domain which contains autophosphorylation sites and is critical for substrate binding. As a kinase, ATM has a serine/threonine kinase domain that is highly homologous to that of the PI3K. At the end of its C-terminus is the FATC domain that is essential for ATM full activation and interactions with partners. Of note, ATM contains multiple important autophosphorylation sites that can substantially affect its functions. For instance, Ser367, Ser1893, Ser1981, and Ser2996 are auto-phosphorylated after irradiation. Thr1885 is not induced by irradiation. In human cells, autophosphorylation on three residues Ser367, Ser1893, Ser1981, and acetylation on Lys3016 have been shown to be important for ATM activation. Aurora B phosphorylates ATM at Ser1403 during mitosis. Importantly, ATM also has 49 α helical HEAT motifs whose name is derived from huntingtin, elongation factor 3, the A subunit of protein phosphatase 2A, and target of rapamycin 1 (TOR1). These motifs serve as scaffolds and are critical for ATM interactions with proteins and DNA.